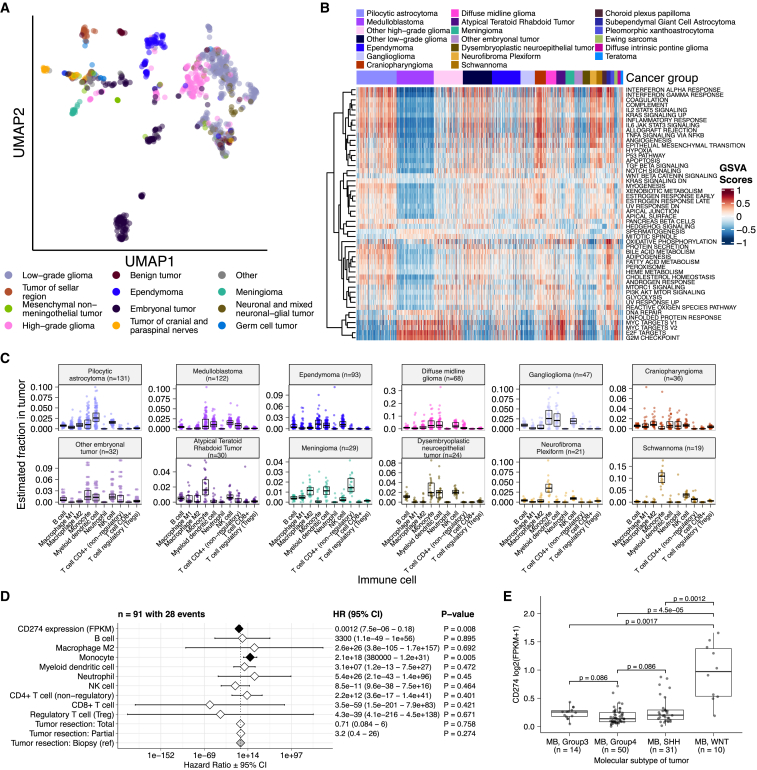
Figure 5.
Transcriptomic and immune landscape of pediatric brain tumors
(A) First two dimensions of transcriptome data UMAP, with points colored by broad histology.
(B) Heatmap of GSVA scores for Hallmark gene sets with tumors ordered by cancer group.
(C) Boxplots of quanTIseq estimates of immune cell proportions in cancer groups with n >15 tumors. Note: other HGGs and other LGGs have immune cell proportions similar to DMG and pilocytic astrocytoma, respectively, and are not shown.
(D) Forest plot depicting additive effects of CD274 expression, immune cell proportion, and extent of tumor resection on OS of medulloblastoma patients. HRs with 95% confidence intervals and p values (multivariate Cox) are listed. Black diamonds denote significant p values, and gray diamonds denote reference groups. Note: the macrophage M1 HR was 0 (coefficient = −9.90e4) with infinite upper and lower confidence intervals (CIs) and thus was not included in the figure.
(E) Boxplot of CD274 expression (log2 FPKM) for medulloblastomas grouped by subtype. Bonferroni-corrected p values from Wilcoxon tests are shown. Boxplot represents 5% (lower whisker), 25% (lower box), 50% (median), 75% (upper box), and 95% (upper whisker) quantiles. Only stranded RNA-seq data are plotted.