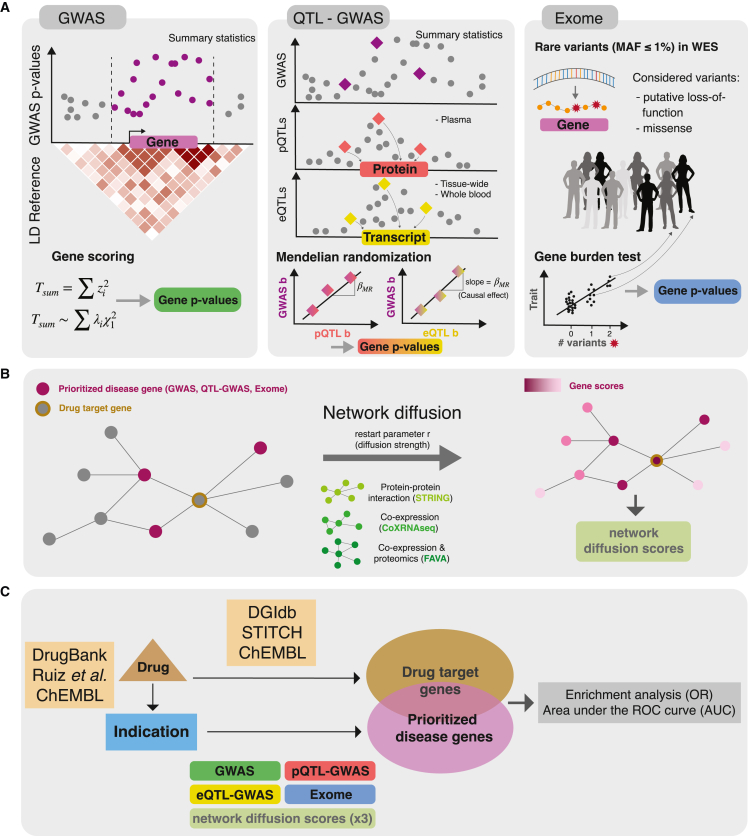
Figure 1.
Overview of the analysis workflow
(A) Three different gene prioritization methods were tested in this study. The first one uses GWAS summary statistics as input (GWAS). The second combines molecular QTL and GWAS summary statistics (QTL-GWAS): either expression QTL (eQTL) or protein QTL (pQTL) data. The third leverages individual-level whole-exome sequencing (WES) data (Exome). In the GWAS method, gene p values are based on the sum of squared SNP Z scores () that follows a weighted distribution. The QTL-GWAS method integrates QTL and GWAS summary statistics through Mendelian randomization (MR). MR causal effect sizes () are calculated from GWAS and mQTL effect sizes (GWAS b and mQTL b, respectively) and gene scores are the corresponding p values. The Exome method aggregates rare variants from WES data. Putative loss-of-function and missense variants with minor allele frequencies (MAF) below 1% are collapsed in burden tests, which results in gene p values. The different approaches were benchmarked for their ability to prioritize drug target genes.
(B) The effects of network diffusion using three different network types and different diffusion strengths (i.e., restart parameter r) were evaluated. Drug target genes may be prioritized only following signal propagation from neighboring disease genes.
(C) Diseases were linked to target genes through public drug databases: first, we used drug-indication information to connect the 30 traits to drugs and then leveraged drug target information to link the drugs to genes. Prioritized disease genes and corresponding diffusion scores (obtained via strategies described in A and B) were then tested for overlap with drug target genes through Fisher’s exact test, resulting in odds ratios (ORs), and through area under the receiver operating characteristic curve (AUC) values.