Figure 3.
Enrichment of prioritized genes for drug targets
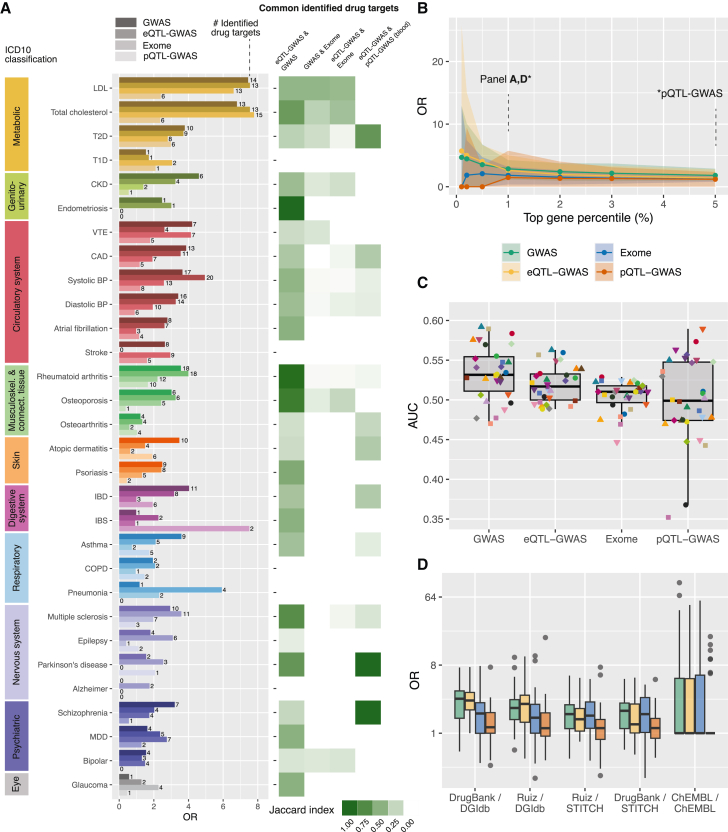
(A) Left: bar plot with odds ratios (ORs) calculated from Fisher’s exact tests between drug target genes and the top 1% (5% for pQTL-GWAS) prioritized genes for the four tested methods and 30 traits, classified according to trait category. Drug target genes were defined by DrugBank/DGIdb, and only drug target genes that could be tested by the respective method were considered. The number on the right of each bar indicates the number of identified drug target genes. Right: overlap of identified drug target genes between pairs of methods quantified through the Jaccard index. The blood-only eQTL-GWAS gene prioritization method was used for the comparison with the pQTL-GWAS method. Plots using UKBB GWASs only are shown in Figure S3.
(B) ORs at different top prioritized gene percentiles for the four methods. The plotted dots correspond to the median OR across the 30 traits, and the shaded area bounds the 10% and 90% percentiles.
(C) Boxplots showing the area under the receiver operating characteristic curve (AUC) values. AUC values were calculated for each trait as indicated by the points (legend in Figure 2) and using the same background genes and drug target definitions as in (A).
(D) ORs calculated for the five drug target definitions and for all four methods (legend in B). The OR was set to 1 for traits with no identified drug target genes. In (C) and (D), the boxplots bound the 25th, 50th (median, center), and 75th quantiles. Whiskers range from minima (Q1 − (1.5 × IQR)) to maxima (Q3 + (1.5 × IQR)) with points outside representing potential outliers.