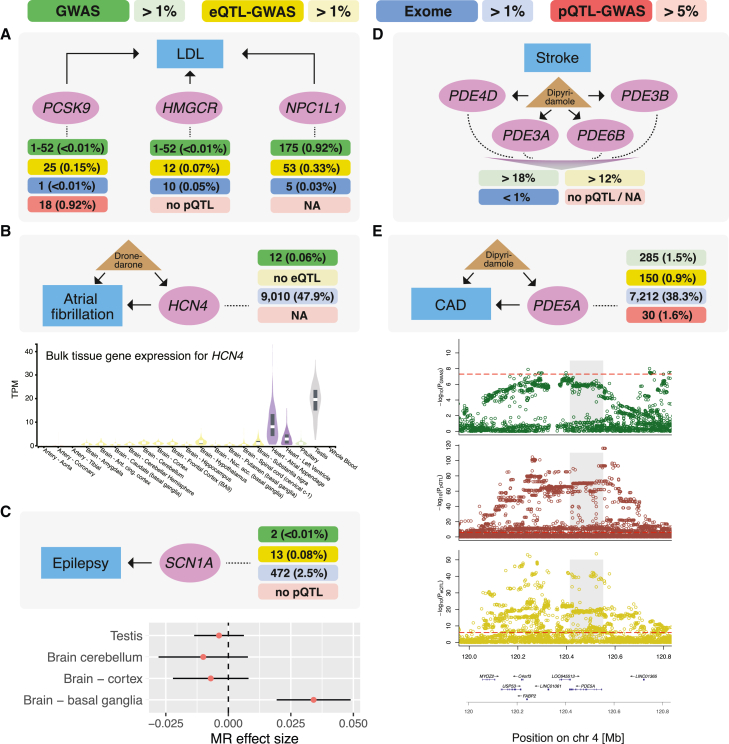
Figure 4.
Examples illustrating drug target genes and their prioritization ranks
(A) Three drug target genes (PCSK9 [evolocumab, alirocumab], HMGCR [statins], and NPC1L1 [ezetimibe] shown in purple) for LDL cholesterol (blue box) and their prioritization ranks (top percentiles shown in parentheses) of each of the four methods (GWAS in green, eQTL-GWAS in yellow, Exome in blue, and pQTL-GWAS in red). Genes that were not testable by a given method are reported as NA (no e/pQTL means that the gene was measured, but had no QTL), and a range of ranks (i.e., 1–52) indicates tied p values.
(B) Top plot shows the prioritization ranks of HCN4, the target of the antiarrhythmic drug dronedarone. Bottom plot shows the gene expression profile of HCN4 across GTEx tissues (TPM, transcripts per million) with “testis,” “heart-atrial appendage,” and “heart-left ventricle” dominating.
(C) Top plot shows the prioritization ranks of SCN1A (sodium voltage-gated channel alpha subunit 1), a drug target gene of several antiepileptic drugs. Bottom plot shows Mendelian randomization (MR) effects (red dots) with 95% CI (black bars) across tissues in which there was a significant eQTL.
(D) Antiplatelet drug dipyrimadole and gene prioritization ranks of its multiple drug targets (a non-exhaustive selection) of the phosphodiesterase (PDE) superfamily.
(E) Top plot shows the gene prioritization ranks of PDE5A, another reported target for dipyrimadole. Bottom plot shows the regional SNP associations (−log10(p)) with coronary artery disease (CAD; GWAS, green), PDE5A protein (pQTL, red), and PDE5A transcript (eQTL, yellow) (red dashed lines indicate the significance thresholds of the respective SNP association, and gray shading marks the position of PDE5A). Bottom row illustrates the position and strand direction of the genes in the locus.