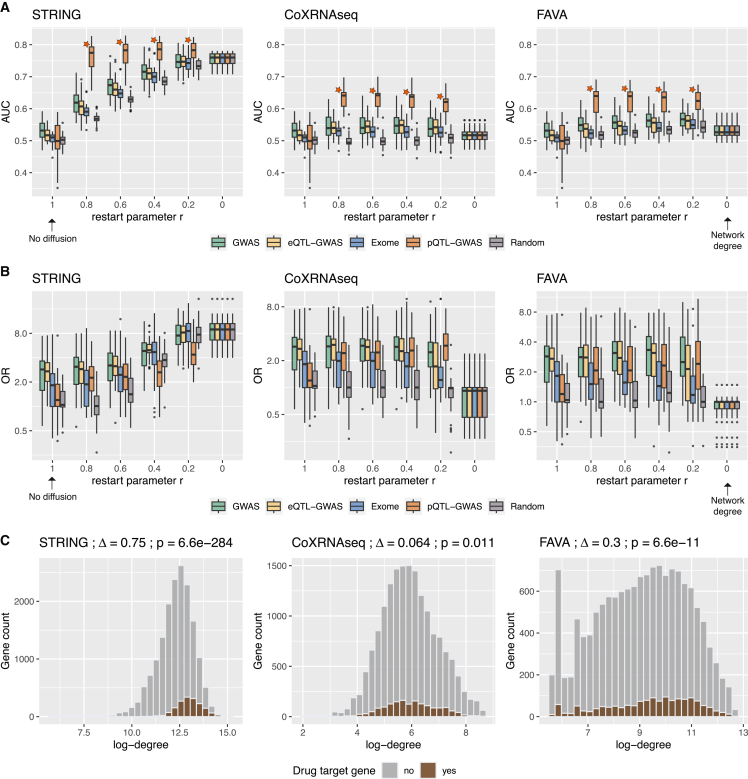
Figure 5.
Effect of network diffusion to prioritize drug target genes
(A) Boxplots showing the area under the receiver operating characteristic curve (AUC) values for each network type (STRING, CoXRNAseq, and FAVA) and method at different restart parameter values r. AUC values were calculated for each of the 30 traits, and drug target genes were defined by DrugBank/DGIdb. At an r value of 1 (no network diffusion), the analysis corresponds to the results in Figure 3B, and at an r value of 0, the gene prioritization rank is based simply on the degree of the network nodes. At 1, the background genes are the genes reported in the respective network. The star next to the pQTL-GWAS method signals that the set of testable genes for this method is enriched for drug target genes, and therefore, higher AUC values were obtained when adding background genes with zero-valued initial scores.
(B) Odds ratios (ORs) between prioritized genes (top 1%) and drug target genes for each network type and method at different r values across the 30 traits (same drug target and background genes as in A). The OR was set to 1 for traits with no identified drug target genes.
(C) Histograms showing the degree distribution of drug target genes and non-drug target genes in each network. The difference in log-degree () and the p values from two-sided t tests are shown at the top. In (A) and (B), the boxplots bound the 25th, 50th (median, center), and 75th quantile. Whiskers range from minima (Q1 − (1.5 × IQR)) to maxima (Q3 + (1.5 × IQR)) with points above or below representing potential outliers.