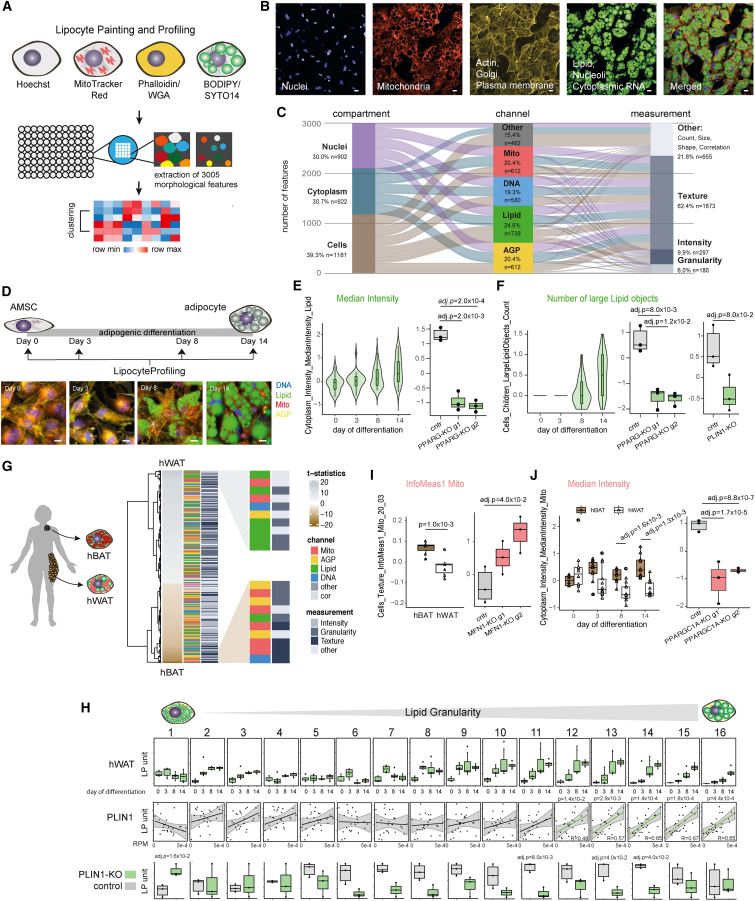
Figure 1.
LipocyteProfiler creates rich morphological and cellular profiles in adipocytes that are informative for known cellular functions
(A) Schematic of LipocyteProfiler, which is a high-content imaging assay that multiplexes six fluorescent stains imaged in four channels in conjunction with an automated image-analysis pipeline to generate rich morphological and cellular profiles in lipid-storing cell types (lipocytes), such as adipocytes during differentiation.
(B) Representative microscopy image of fully differentiated adipocytes for four individual channels and a merged representation across channels. Scale bars, 10 μm.
(C) LipocyteProfiler extracts 3,005 morphological and cellular features that map to three cellular compartments and across four channels using four measurement classes.
(D) Schematic of LipocyteProfiling in differentiating hWAT at four time points of adipocyte differentiation (days 0, 3, 8, 14). Representative images of AMSCs stained using LipocytePainting at four time points of differentiation (days 0, 3, 8, 14). Scale bars, 10 μm.
(E) Cytoplasm MedianIntensity Lipid, a measurement of lipid content within a cell, significantly increases with adipogenic differentiation and decreases following CRISPR-Cas9-mediated knockdown of PPARG in differentiated white adipocytes. Data are shown for two guides used (g1 and g2), and y axis shows LP units (normalized LipocyteProfiling [LP] values across three batches, see STAR Methods).
(F) Number of large Lipid objects informative for large lipid droplets are absent in the progenitor state (day 0) and in early differentiation (day 3) and progressively increase in later stages of differentiation (days 8 and 14). Number of large Lipid objects is reduced following CRISPR-Cas9-mediated knockout (KO) of PPARG (data are shown for two guides used [g1 and g2]) and PLIN1, at day 14 of differentiation. y axis shows LP units (normalized LP values across three batches, see STAR Methods).
(G) Morphological profiles of white (hWAT) and brown (hBAT) adipocytes at day 14 of differentiation differ significantly across all feature classes (FDR < 0.1%). Features are clustered based on effect size. Features with the highest effect size in hWAT and hBAT adipocytes are lipid- and mitochondria-related, respectively. Graph shows zoom-in for top ten features with largest effect sizes in hWAT (top panel) and hBAT (bottom panel).
(H) Lipid Granularity measures, as spectra of 16 lipid-droplet size measures, show size-specific changes in hWAT and hBAT during differentiation. See also Figure S1H. Granularity features informative for larger lipid droplets (Lipid Granularity 10–16) correlate positively with PLIN1 gene expression and are reduced in PLIN1-KO adipocytes. See also Figures S1I and S1J (PLIN2, FASN-KO). y axis shows autoscaled LP units (normalized LP values across three batches, seeSTAR Methods).
(I) Brown adipocytes (hBAT) show higher Mito_Texture_InfoMeas1, a measure of spatial relationship between specific intensity values, compared with white adipocytes (hWAT). CRISPR-Cas9-mediated knockout of MFN1, a mitochondrial fusion gene, changes Mito_Texture_InfoMeas1 (data shown for two guides used [g1 and g2]). y axis shows LP units (normalized LP values across three batches [hBAT/hWAT] or normalized across CRISPR-KO data, see STAR Methods).
(J) Mito_MedianIntensity is higher in brown (hBAT) compared with white (hWAT) adipocytes throughout differentiation and decreased after CRISPR-Cas9-mediated knockout of PPARGC1A in hWAT. y axis shows LP units (normalized LP values across three batches, see STAR Methods).