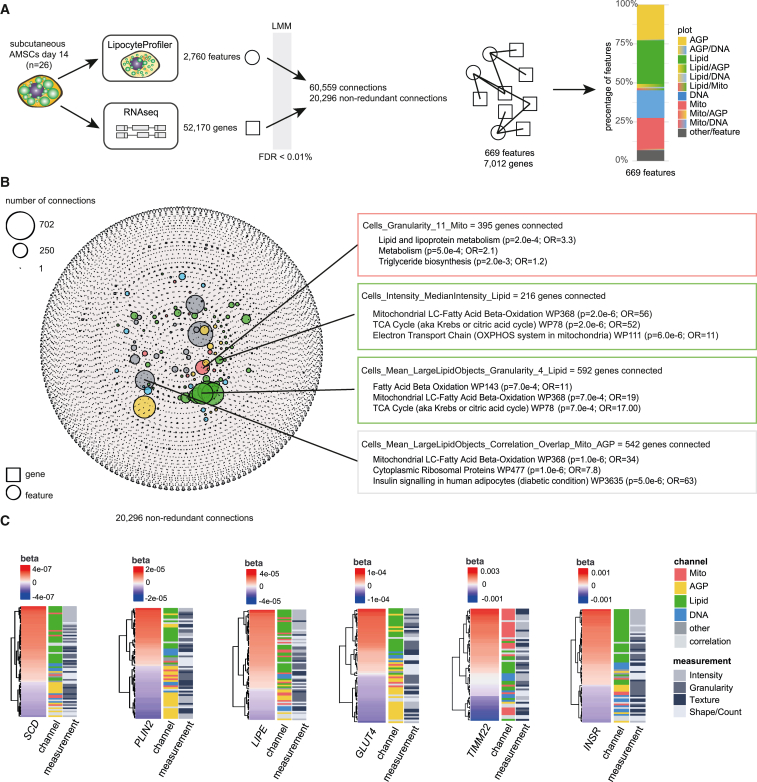
Figure 3.
Correlations between morphological and transcriptional profiles
(A) Linear mixed model (LMM) was applied to correlate 2,760 morphological features derived from LipocyteProfiler with 52,170 transcripts derived from RNA-seq in matched samples of subcutaneous AMSCs at terminal differentiation (day 14). With FDR < 0.01%, we discover 20,296 non-redundant connections that map to 669 morphological features and 7,012 genes.
(B) Network of transcript-LipocyteProfiler feature correlations (significant connections FDR < 0.01%). Genes correlated with individual LipocyteProfiler features are enriched for relevant pathways (FDR < 5%). Node size is determined by number of connections. See also Figure S3A for a network with a significance level threshold of FDR < 0.1%.
(C) LipocyteProfiler signatures of adipocyte marker genes SCD, PLIN2, LIPE, GLUT4, TIMM22, and INSR recapitulate their known cellular function. Features are clustered based on beta of linear regression.