Figure 1. p53 negatively regulates ATX expression in MEF.
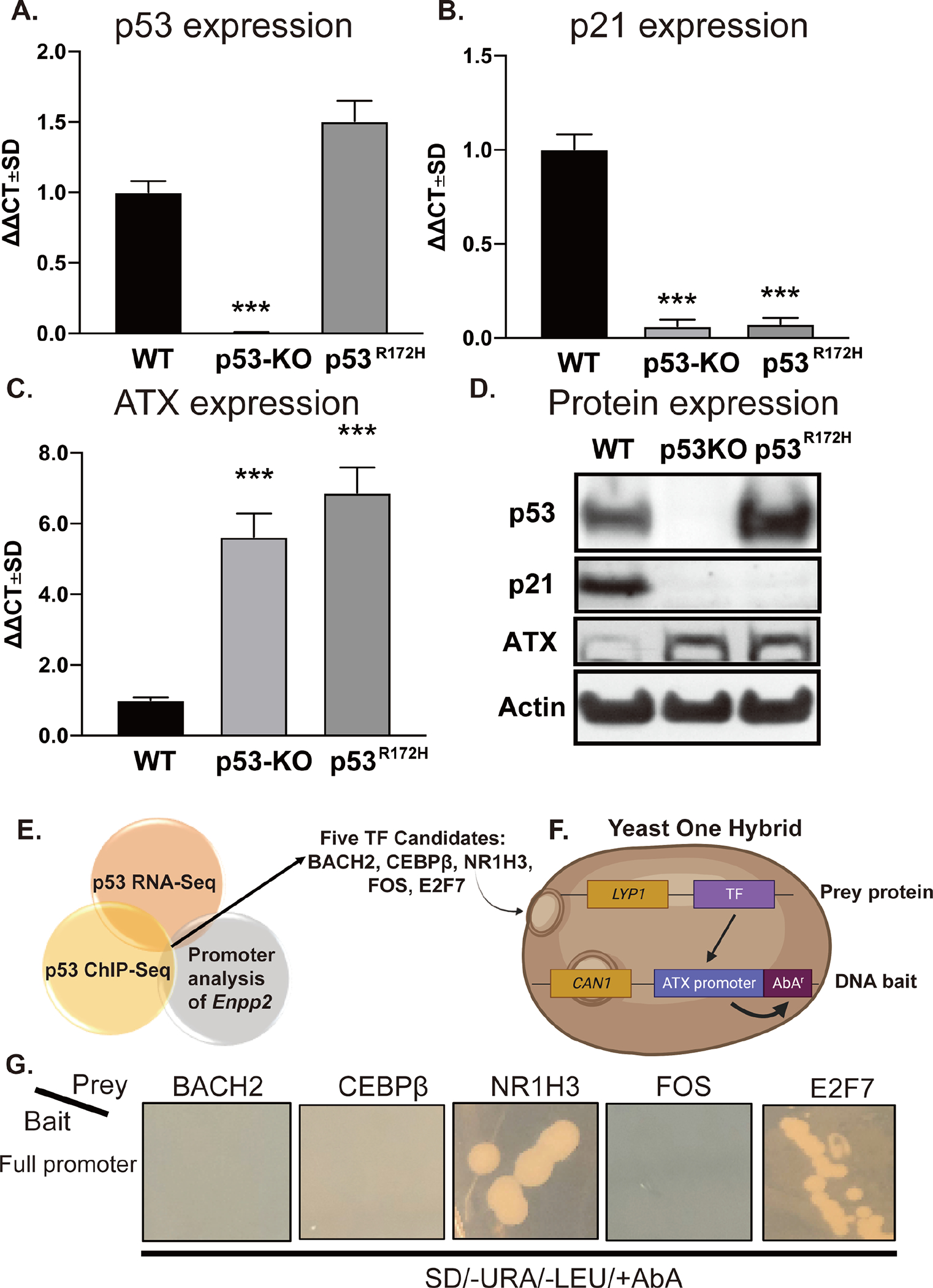
RNA was isolated from WT, p53-KO, and p53R172H MEF and p53 (A), p21 (B), and ATX (C) mRNA expression was evaluated by RT-qPCR. Fold changes in mRNA (ΔΔCt) were calculated, and data are represented as the mean ± SD of three independent experiments. *** indicates p < 0.001. (D) Expression of p53, p21, and ATX were determined by immunoblotting with antibodies in cell lysates prepared from WT, p53-KO, and p53R172H MEF. Note the robust upregulation of ATX protein in p53-KO and p53R172H MEF. (E) The results of the promoter analysis of Enpp2 were cross-referenced with published p53 ChIP-Seq and RNA-Seq databases. The dissimilarity threshold was set at 10% as the selection criterion. (F) Five TFs, BACH2, CEBPβ, NR1H3, FOS, and E2F7, were identified and cloned as “prey protein” into the pGLDT7-AD vector. The Enpp2 promoter sequence was cloned to “DNA bait” in pABAi vector, which contains an AbA resistance gene as described in the methods. (G) The pairs of prey and bait vectors were transformed to Y1HGold yeasts and grown on plates in the presence of SD-Leu-URA+AbA. The positive interaction between the prey protein and the DNA bait caused the expression of AbAr and growth in the plate. LYP1: lysine permease; CAN1: plasma membrane arginine permease; SD: synthetic defined medium.
