Figure 3. E2F7 enhances the transcriptional activity of the ATX promoter.
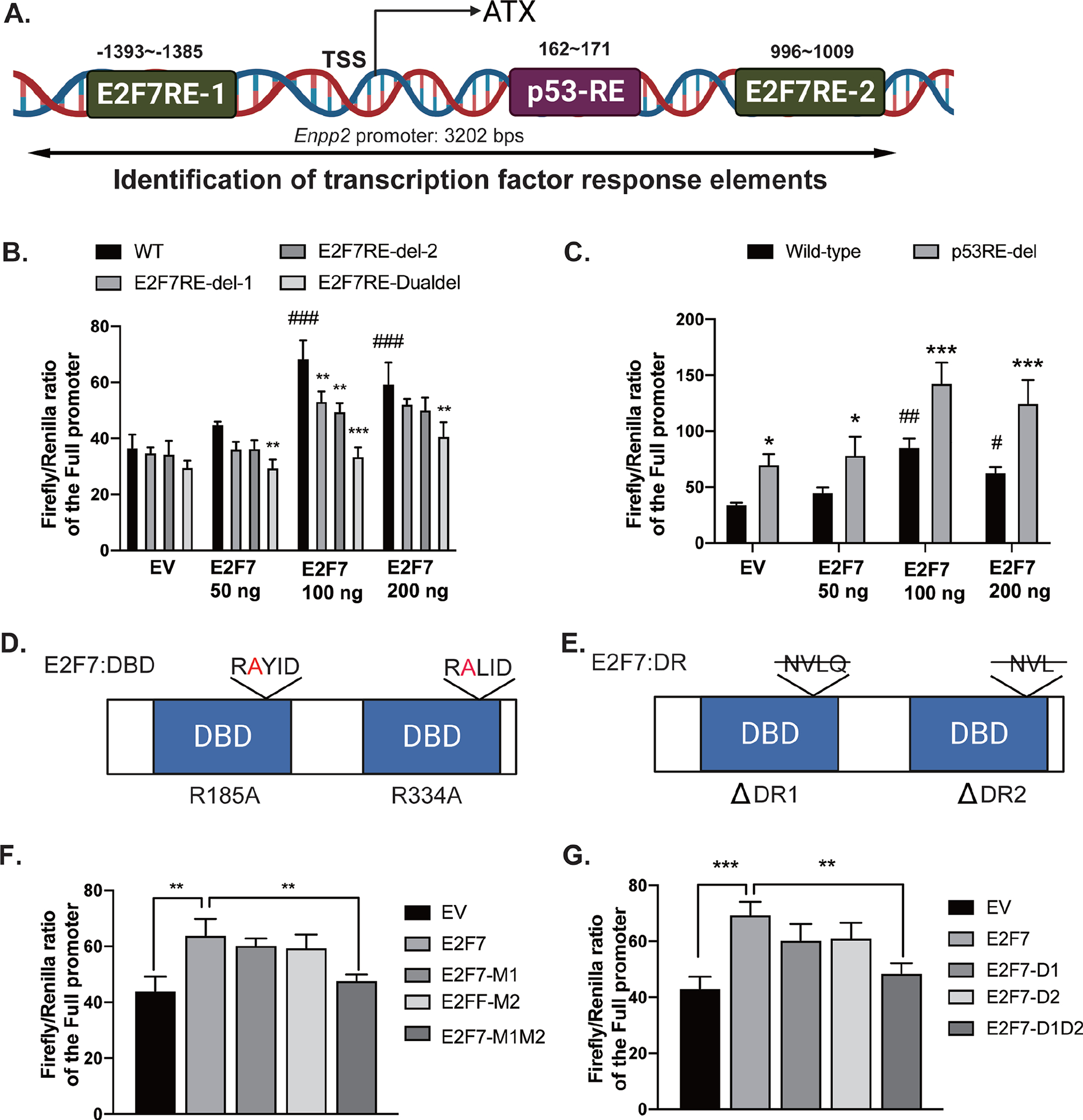
(A) A 3202 bp Enpp2 promoter was retrieved from Ensembl database and analyzed by TRANSFAC and ALGGEN-PROMO online software. Two potential E2F7-RE (−1393~−1385, CCCGCTCTG, E2F7-RE1; 996~1009, TGTCTCCCCGGGAA, E2F7-RE2) and one p53-RE (162~171, TTGGCAGGAT) were identified. (B) Single deletion (E2F7RE-del-1 or E2F7RE-del-2) or double deletion (E2F7RE-Dualdel) of E2F7-RE and (C) deletion of p53-RE were introduced into the reporter plasmids. Co-transfection of E2F7-expression vector and luciferase reporter was carried out in HEK293T cells as described in methods. (D-E) The diagrams show mutations in the DNA binding domain (DBD) and dimerization residues (DR) of E2F7, respectively. Single arginine-to-alanine mutation (M1 or M2) or double mutations (M1M2) of E2F7 were generated in the E2F7-expression vector. Likewise, single deletion of DR (D1 or D2) and double deletions (D1D2) were generated in the E2F7-expression vector. (F) The E2F7-DBD mutation and (G) the E2F7-DR mutation vectors were co-transfected with Enpp2 promoter reporter plasmid into HEK cells. Luciferase activity was measured after transient co-transfection of these plasmids. Luciferase activity was normalized to Renilla luciferase activity. Data are represented as the mean ± SD of three independent experiments. *, #p < 0.05, **, ##p<0.01, and ***, ###p < 0.001 indicate significant differences. * indicates the comparison between the values from wild-type promoter and E2F7-RE deletion promoter plasmids. # indicates the comparison between EV and E2F7-expression vectors. EV: empty vector; DBD: DNA binding domain; DR: dimerization residue.
