FIGURE 3.
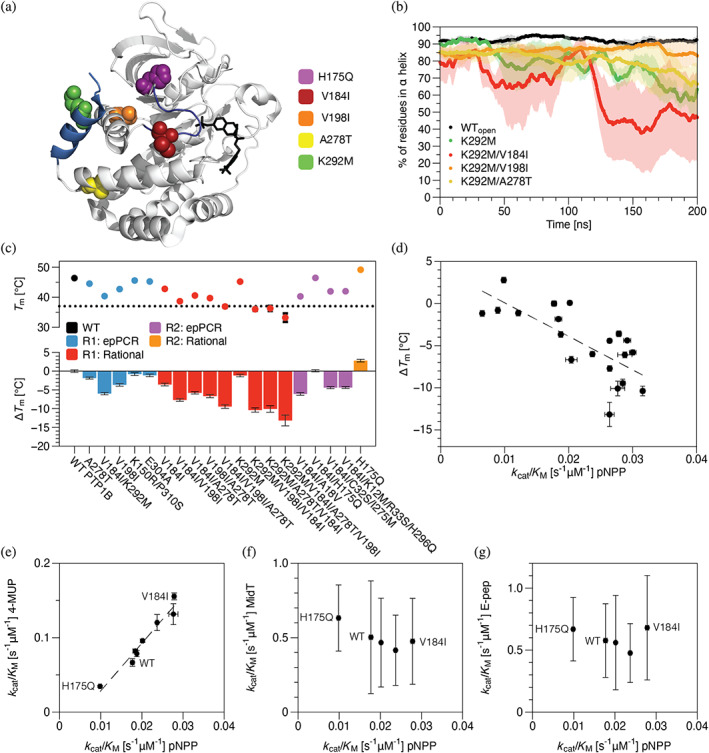
Mutations identified in neutral drift tend to reduce thermodynamic stability but exhibit substrate‐specific effects on activity. (a) A crystal structure of PTP1B (PDB entry 2f71) shows the locations of a subset of neutral (V198I and A278T), activating (V184I), or inactivating (K292M and H175Q) mutations identified in neutral drift experiments (PDB entry 2a5j). Highlights: competitive inhibitor (black), ⍺7 helix (285–298, light blue), and WPD loop (178–185, dark blue). (b) The percent of residues in the core of the ⍺7 helix (287–295) that exhibit ⍺‐helical conformations in each uncorrelated trajectory frame of MD trajectories. Mutant K292M accelerates ⍺7 helix disordering, and V198I suppresses this disruptive effect. (c) We used differential scanning calorimetry (DSF) to measure changes in the melting temperature of PTP1B: ΔT m = T m‐mut − T m‐WT. Most mutations reduced the melting temperature, an effect that was largely additive when mutations were combined. (d) Mutation‐derived shifts in melting temperature and catalytic activity are moderately correlated (r 2 = 0.55). (e) The activities of mutants on two model substrates—pNPP and 4‐methylumbelliferyl phosphate (4‐MUP)—are strongly correlated and differ by two‐ to four‐fold, depending on substrate; all have similar activities on (f) MidT and (g) E‐Pep, two phosphopeptides. MidT = EPQpYEEIPIYL, and E‐Pep = DADEpYLIPQQG. In b‐g, data depict the mean and standard error for n ≥ 3 technical replicates (MD and kinetic data) or mean and standard deviation for n ≥ 12 technical replicates (T m).
