FIGURE 7.
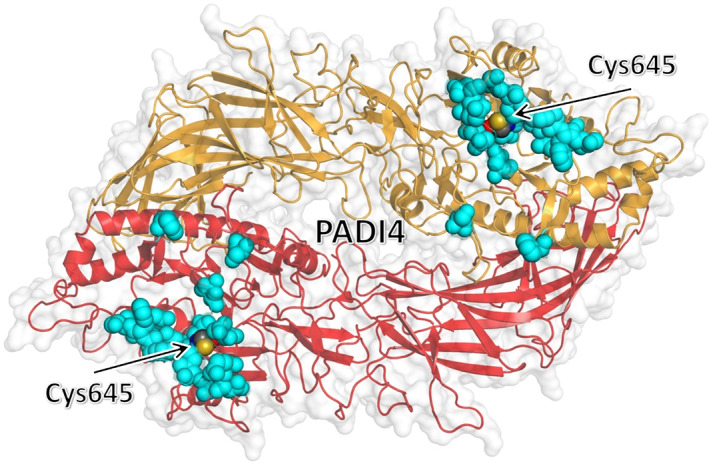
Clustering of residues of PADI4 with the most favorable affinity toward the p53‐binding domain of N‐MDM2. Top three residues of PADI4 with the most favorable binding affinity to each of the eight selected docking poses, as obtained by using MM/GBSA with per residue decomposition (Table 1), are shown (cyan) in van der Waals representation. The key catalytic residue Cys645 is also explicitly shown and labeled. The two chains of the homodimeric structure of PADI4 are represented in different colors.
