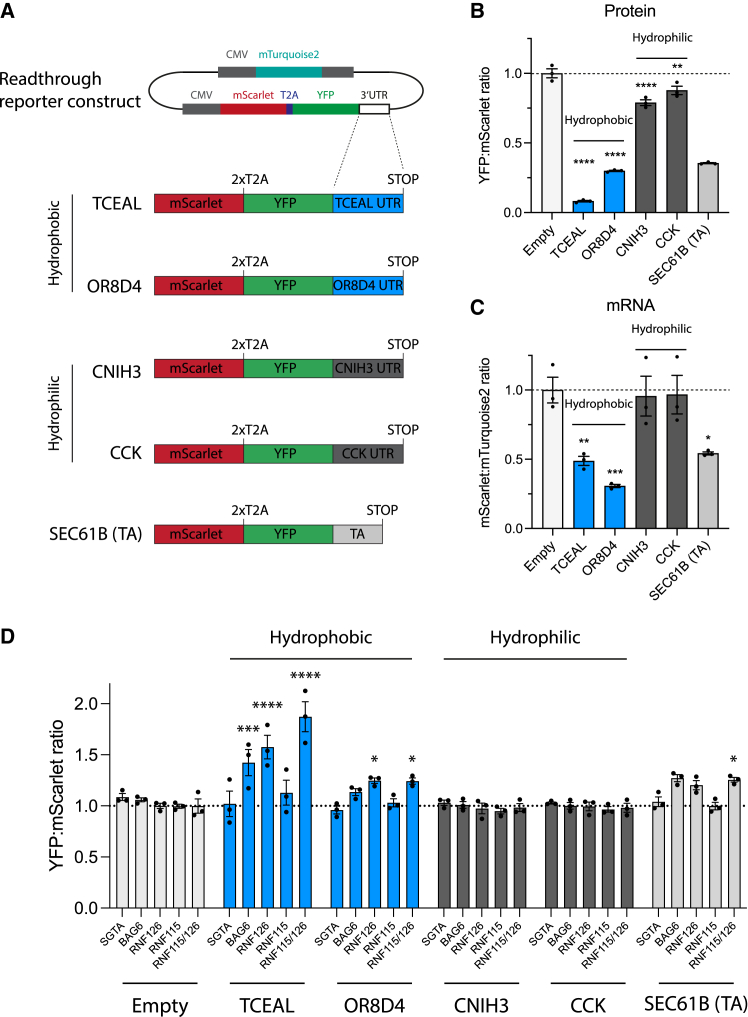
Figure 4.
Readthrough mitigation pathways are conserved in mammalian cells
(A) Constructs for ratiometric analysis by flow cytometry of effects of readthrough into 3′ UTRs encoding hydrophobic (transcription elongation factor A protein-like 1 [TCEAL1], KDS = 2.07, 32 residues; olfactory receptor 8D4 [OR8D4], KDS = 1.96, 28 residues) or hydrophilic (protein cornichon homolog 3 [CNIH3], KDS = −2.21, 34 residues; cholecystokinin [CCK], KDS = −1.89, 26 residues) CTE sequences in HEK293T cells. The TA sequence of SEC61B was also analyzed.
(B and C) Ratiometric analysis in HEK293T cells of protein levels (YFP:mScarlet ratio) (B) and of mRNA levels (mScarlet:mTurquoise2 ratio) (C) of constructs in (A). Data from flow cytometry (see Figure S4C). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by Dunnett’s test. Error bars represent mean ± SEM (n = 3). Dotted line indicates empty control ratios.
(D) Effects of the deletion of genes encoding factors involved in readthrough mitigation on protein levels of hydrophilic and hydrophobic readthrough constructs, determined as in (B). Error bars represent mean ± SEM (n = 3). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 by Dunnett’s test. Dotted line indicates wild-type ratios.