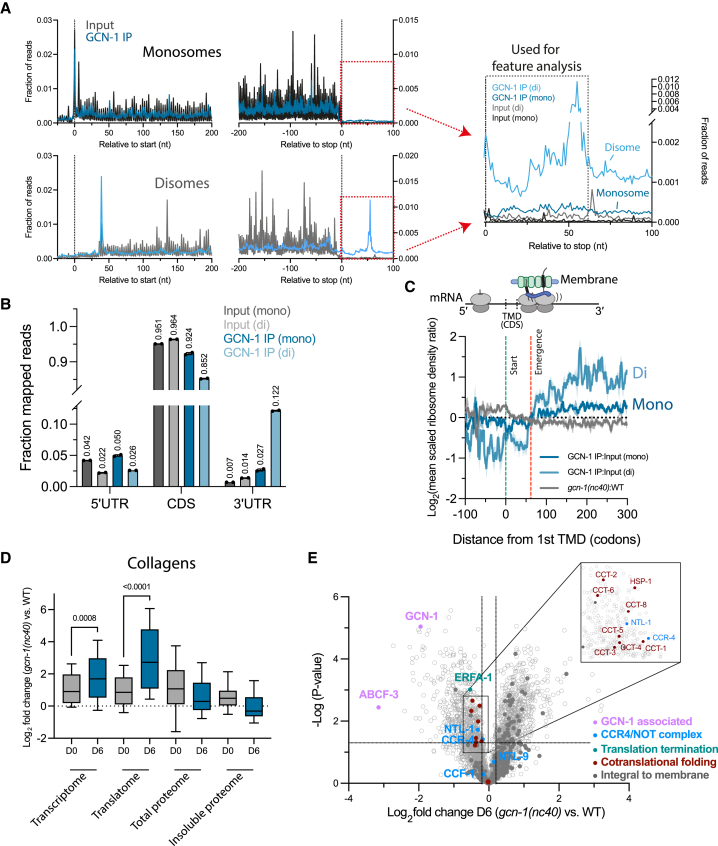
Figure 5.
Selective ribosome profiling reveals GCN-1 binding to hydrophobic ribosomes translating 3′ UTRs, TMD proteins, and collagens
(A) Metagene plots of GCN-1-bound ribosomes (monosomes and disomes) and total input control are shown (3′ UTR regions magnified in right) (see STAR Methods).
(B) Distribution of RPFs of GCN-1-IPed ribosomes and total input control in 5′ UTR, coding sequences (CDSs) and 3′ UTR regions of monosomes and disomes. Mean values are indicated above bars.
(C) Metagene analysis of GCN-1 interaction with TMD protein transcripts. RPFs of GCN-1 bound monosomes (odds ratio compared with total input; n = 1,303; blue line ± SEM in light blue), disomes (n = 2,595; light blue line ± SEM shaded) and ribosomes of gcn-1(nc40) mutant animals (odds ratio compared with wild type; n = 1,029; dark gray line ± SEM in light gray) are shown. Each transcript was centered around the onset of the first TMD (position 0, green dotted line) and RPFs were expressed as mean-scaled ribosome densities (normalization window of 300 codons up- and downstream of TMD start, position 0). Full emergence of TMDs from the ribosome exit tunnel is indicated by the red dotted line at codon position 65, assuming an average TMD length of 25 codons and a ribosomal exit tunnel length of 40 codons.
(D) Age-dependent effects of GCN-1 dysfunction on the transcriptome (n = 130), translatome (n = 110), total proteome (n = 39), and insoluble proteome (n = 29) of collagens. Young (day 0) and old (day 6) gcn-1(nc40) mutant worms were analyzed relative to young (day 0) and old (day 6) wild-type nematodes, respectively. The horizontal line within boxplots indicates the median; boxes indicate upper and lower quartile and whisker caps 10th–90th percentile, respectively. p values by Holm-Sidak test. See also Tables S1E–S1H, S2A, S2B, S3A, and S3B.
(E) Volcano plot representation of label-free proteome analysis of polysome fractions of old (day 6) gcn-1(nc40) mutant worms relative to polysome fractions of day 6 wild-type animals (as in Figure 3G). Selected proteins are annotated. Dotted lines indicate cutoffs for enrichment at the x axis (log2 ± 0.2, ∼1.15-fold) and at the y axis for p values (0.05, −log > 1.33). See also Figure S3F and Table S1I.