Figure S3.
Effects of 3′ UTR translation on mRNA and protein levels, related to Figure 3
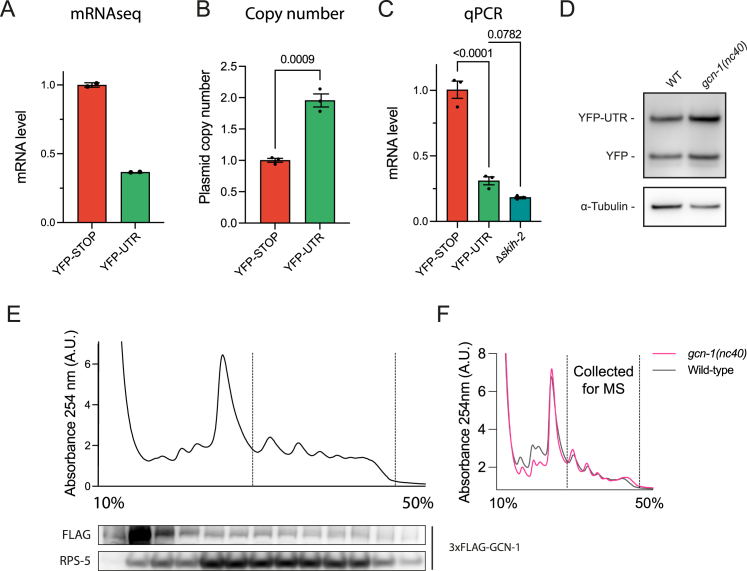
(A) mRNA-seq analysis of C. elegans reporter strains expressing YFP-UTR and YFP-STOP (n = 2). Error bars represent mean ± SEM.
(B) qPCR analysis of copy number of YFP-UTR and YFP-STOP in C. elegans reporter strains. Integrated gene copy numbers are on average 1.96 ± 0.11-fold higher in YFP-UTR compared with YFP-STOP. Error bars represent mean ± SEM (n = 3). Data were analyzed using the 2(−ΔΔCt) formula. p values by unpaired t test.
(C) qPCR analysis of YFP-STOP and YFP-UTR mRNA levels in wild-type C. elegans and in skih-2 mutant animals. Error bars represent mean ± SEM (n = 3). Data were analyzed using the 2(−ΔΔCt) formula. p values by Fisher’s LSD test.
(D) Representative immunoblot analysis of wild-type or gcn-1(nc40) mutant worms expressing YFP-UTR. See Figure 3C for quantification.
(E) Representative trace of sucrose density gradient fractionation (A254 nm; top) and immunoblot analysis of 3x-FLAG-tagged GCN-1 in C. elegans. Dotted lines delineate polysome fractions.
(F) Representative traces of sucrose gradient density fractionation of wild-type and gcn-1(nc40) mutant animals (A254 nm). Dotted line indicates polysomes collected for subsequent MS/MS analysis shown in Figures 3G and 5E.