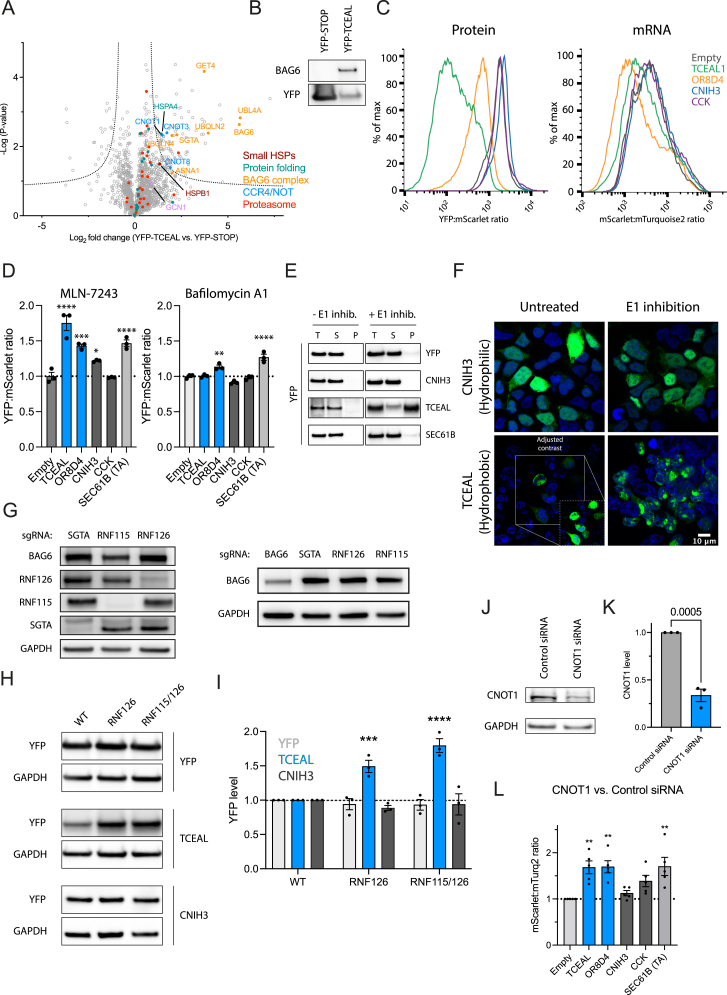
Figure S4.
Protein and mRNA clearance upon translational readthrough is conserved in HEK293T cells, related to Figure 4
(A) Volcano plot representation of label-free interactome analysis of YFP-TCEAL from HEK293T cells expressing YFP-TCEAL CTE construct and YFP-STOP as control. Components of the BAG6, proteasomal subunits, heat-shock proteins (e.g., HSPB1 and HSPA4) and CCR4/NOT complexes are identified as interactors of YFP-TCEAL. Selected proteins are highlighted. See also Table S1L.
(B) Immunoprecipitation of YFP-STOP or YFP-TCEAL with anti-GFP antibody from HEK293T cells expressing the respective constructs. Fractions were analyzed by immunoblotting for BAG6 and YFP (n = 2). Note that the UTR sequence adds 30 amino acids (mainly hydrophobic) to the C terminus of YFP, resulting in only slightly slower migration of YFP-TCEAL compared with YFP-STOP.
(C) Representative histograms of flow cytometry analysis indicating YFP:mScarlet and mScarlet:mTurquoise2 ratios of cells transiently transfected with the indicated reporter plasmids (related to Figures 4B–4D).
(D) Ratiometric flow cytometry analysis of cells expressing the indicated reporter plasmids (see Figure 4A) in the presence of the E1 ubiquitin-activating inhibitor MLN-7243 (compared with untreated cells) (n = 3; left) and the lysosomal degradation inhibitor bafilomycin A1 (compared with untreated cells) (n = 3; right). Error bars represent mean ± SEM. p values by Dunnett’s test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
(E) Representative immunoblot analysis of soluble and pellet fractions from cells expressing the indicated reporter constructs (see Figure 4A). YFP-TCEAL is recovered in the pellet fraction upon E1 inhibition.
(F) Representative fluorescence microscopy images of hydrophobic (YFP-TCEAL) or hydrophilic (YFP-CNIH3) readthrough reporter proteins with or without E1 inhibition by MLN-7243. Insert in the lower left image shows cells after contrast adjustment for the low expression level of the YFP-TCEAL readthrough reporter.
(G) Representative immunoblot analysis of HEK293T depletion cell lines for components of the BAG6 complex.
(H) Representative immunoblot analysis of reporter constructs in different BAG6 mutant backgrounds shown in (F). Related to Figure 4D.
(I) Quantification of immunoblot analysis in (G) by densitometry (n = 3). p value by Dunnett’s test. Error bars represent mean ± SEM. ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Related to Figure 4D.
(J) Representative immunoblot analysis of downregulation efficiency using siRNA against CNOT1 compared with control siRNA. p value by unpaired t test.
(K) Quantification of immunoblot analysis in (I) by densitometry. Error bars represent mean ± SEM (n = 3). p value by Dunnett’s test.
(L) Effect of downregulation of CNOT1 on mRNA level of 3′ UTR reporter constructs and SEC61B (TA). Ratiometric analysis by flow cytometry of cells treated with siRNA against CNOT1 or control siRNA. Error bars represent mean ± SEM (n = 5). p value by Dunnett’s test. ∗∗∗p < 0.001.