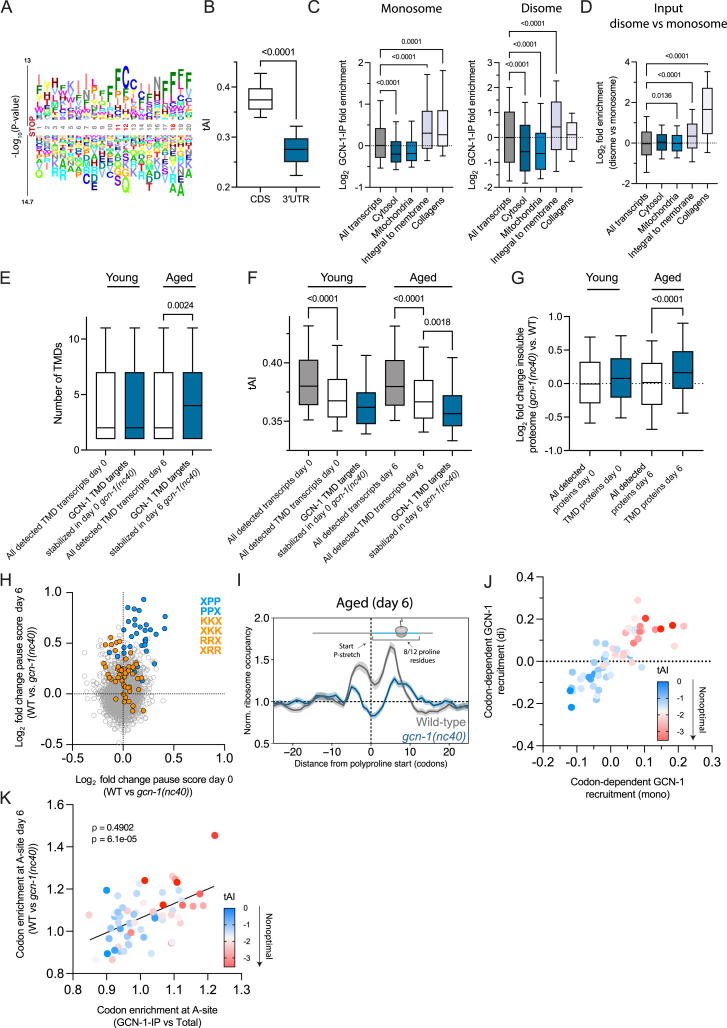
Figure S5.
GCN-1 is recruited to ribosomes translating hydrophobic 3′ UTRs, TMD proteins, and collagens, related to Figure 5
(A) Logo plots of amino acid enrichment using kplogo analysis (Wu and Bartel124). Sequences (20 aa downstream of annotated stop codon) chosen for analysis were derived from 3′ UTRs bound by GCN-1 (monosomes and disomes; in frame 1) indicated in Figure 5A (right). Numbers indicate the position after the stop codon. The y axis indicates the sum of log p values for each amino acid at a given position. Significantly enriched positions are marked in red. Also see Table S2C.
(B) 3′ UTRs are enriched in nonoptimal codons. tRNA adaptation index (tAI) was analyzed for coding sequences (CDSs) (n = 20,190) and 3′ UTRs (n = 14,434) in C. elegans. The horizontal line in the boxplots indicates the median; boxes indicate upper and lower quartile and whisker caps 10th–90th percentile, respectively. p value by unpaired t test.
(C) GCN-1 recruitment (log2 enrichment) across transcripts of proteins from different cellular compartments based on monosomes (left) (all transcripts, n = 12,894; cytosol, n = 432; mitochondria, n = 301; integral to membrane n = 2,158; collagens, n = 115) or disomes (right) (all transcripts, n = 16,294; cytosol, n = 590; mitochondria, n = 478; integral to membrane n = 3,838; collagens, n = 129). The horizontal line in the boxplots indicates the median; boxes indicate upper and lower quartile and whisker caps 10th–90th percentile, respectively. p values by Dunnett’s test. See Tables S2D and S2E.
(D) Disome enrichment (log2 enrichment) across transcripts of proteins from different cellular compartments (all transcripts, n = 15,036; cytosol, n = 584; mitochondria, n = 479; integral to membrane n = 5,733; collagens, n = 154).
(E) GCN-1 dysfunction preferentially stabilizes mRNA levels of integral membrane proteins with multiple TMDs in aged animals. This analysis depicts the number of TMDs that are encoded by GCN-1 target transcripts. These transcripts were defined as meeting two criteria: (1) having 2-fold enrichment by selective ribosome profiling and (2) displaying ∼1.15-fold (log2 0.2) stabilization in the gcn-1(nc40) mutant animals compared with wild type. The distribution of the TMD number in these GCN-1 target transcripts (blue) were compared with the TMD number in all transcripts (white) detected in our experiments in day-0 and day-6 animals. The horizontal line in the boxplots indicates the median; boxes indicate upper and lower quartile and whisker caps 10th–90th percentile, respectively. p value by Holm-Sidak’s test. Also see Table S3C.
(F) TMD transcripts are enriched in nonoptimal codons. tAI analysis of all TMD transcripts (white), TMD transcripts stabilized in gcn-1(nc40) mutant nematodes (blue) in comparison with all coding sequences (CDSs, gray). Only transcripts that were detected in both mRNA-seq and GCN-1 selective ribosome profiling were considered for analysis matching the same criteria as in (E) for day-0 and day-6 animals (also see Table S3C). The horizontal line in the boxplots indicates the median; boxes indicate upper and lower quartile and whisker caps 10th–90th percentile, respectively. p value by Holm-Sidak’s test. See Table S3C.
(G) Age-dependent effect on TMD protein insolubility in GCN-1-deficient (gcn-1(nc40)) nematodes. TMD proteins identified by mass spectrometry in the insoluble fraction of lysates from young (day 0) and aged (day 6) worms were analyzed. The log2 fold increase in insolubility from day 0 to day 6 is shown. The horizontal line in the boxplots indicates the median; boxes indicate upper and lower quartile and whisker caps 10th–90th percentile, respectively. p value by Dunnett’s test. See also Tables S1E–S1H.
(H) Age-dependent effect of GCN-1 dysfunction on translational pausing at tripeptide motifs. Translational pause scores were analyzed from ribosome profiling data for tripeptide motifs in young (x axis, day 0) and old (y axis, day 6) gcn-1(nc40) mutant animals compared with age-matched wild-type animals (n = 18,612 motifs). PP containing motifs are highlighted in blue and KK or RR motifs in orange. The pause score is calculated as the sum of normalized ribosome densities on each triplet amino acid motif. Only transcripts with at least 10 reads and a minimum length of 100 nt were considered for the analysis.
(I) Normalized ribosome occupancy (mean scaled) centered around onset of polyproline stretch (x = 0; dashed vertical line; with at least 8 out of 12 residues being proline) in aged (n = 192 positions from 142 genes) wild-type (gray) and gcn-1(nc40) (blue) animals. The light gray (wild type) and light blue (gcn-1(nc40)) shaded areas indicate the 95% confidence interval.
(J) Codon recruitment coefficients calculated as the Pearson’s correlation coefficient of GCN-1 recruitment (monosomes [x axis] and disomes [y axis]) and codon frequency of transcripts reveal increased GCN-1 recruitment to nonoptimal codons. Codon optimality is represented as a color gradient from blue (optimal) to red (nonoptimal).
(K) Dysfunction of GCN-1 (in gcn-1(nc40) mutant) results in an age-dependent decrease of ribosome pausing at nonoptimal codons. Codon enrichment at A-site of GCN-1-IPed ribosomes relative to total input in (H) (x axis) is compared with codon enrichment at A-site of wild-type relative to gcn-1(nc40) mutant nematodes. Codon optimality (tAI) is indicated by color-scale (red, low codon optimality; blue, high codon optimality). Statistics by Spearman’s correlation (ρ = 0.4902, p = 6.1e−5). Line represents linear regression fit.