Figure S6.
GCN1 function is conserved in HEK293T cells, related to Figure 6
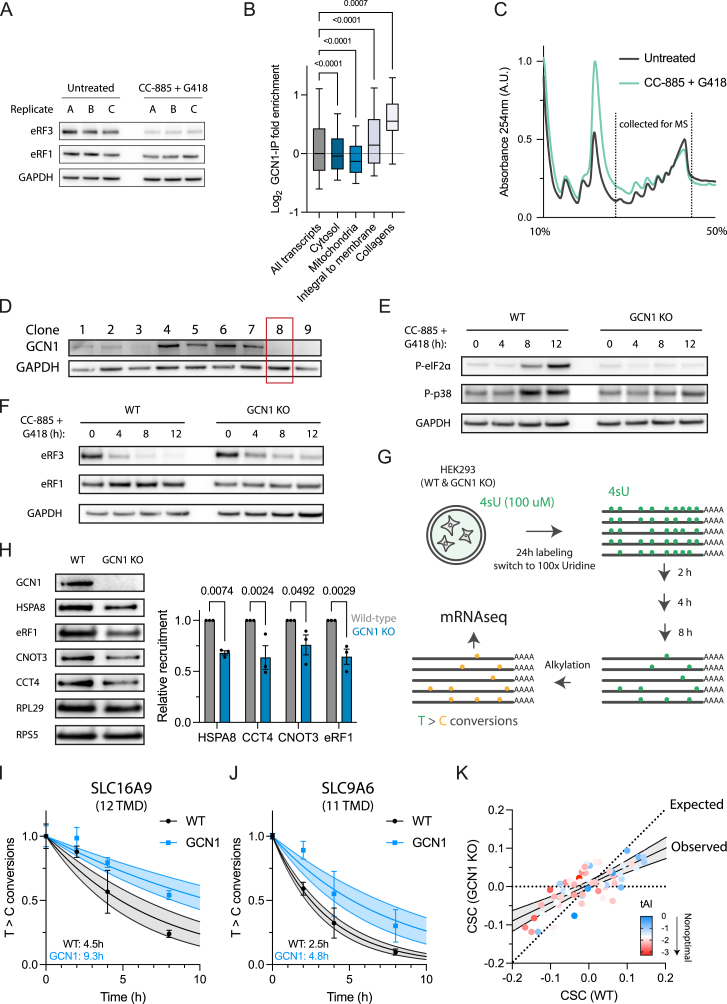
(A) Immunoblot analysis of eRF3 upon treatment of HEK293T cells with CC-885 (10 nM) and G418 (20 μg/mL) for 4 h to induce readthrough (n = 3).
(B) GCN1 recruitment (log2 enrichment) based on selective ribosome profiling in untreated HEK293T cells across transcripts of proteins from different cellular compartments (all transcripts, n = 16,246; cytosol, n = 4,488; mitochondria, n = 1,463; integral to membrane n = 3,301; collagens, n = 60). The horizontal line in the boxplots indicates the median; boxes indicate upper and lower quartile and whisker caps 10th–90th percentile, respectively. p values by Dunnett’s test. See Table S2G.
(C) Sucrose density gradient fractionation of HEK293T cells treated with CC-885 and G418 as in (A). The dotted lines delineate fractions collected for proteome analysis shown in Figure 6C.
(D) Immunoblot analysis of single-cell-sorted polyclonal GCN1 deletion cells. Clone 8 (red square) was chosen for the experiments in Figures 6D, S6D, and S6E.
(E) Representative immunoblot analysis of eIF2α and p38 phosphorylation during eRF3 depletion upon treatment of cells with CC-885 and G418 for the times indicated. Related to Figure 6D.
(F) Representative immunoblot analysis of eRF3 depletion upon treatment of wild-type and GCN1 KO cells with CC-885 and G418 for the times indicated. Related to Figure 6D.
(G) Schematic overview of SLAM-seq workflow. Preexisting mRNAs are labeled for 24 h with 4-thiouridine (4sU). Then the media is exchanged with 100X uridine-containing media, which marks the onset of the chase. Over time, the 4sU-labeled mRNAs will be degraded. The prelabeled mRNA can be distinguished from newly synthesized mRNA by alkylating the 4sU-labeled sites, leading to T > C conversions upon reverse transcription. The loss of T > C conversions over time allows the calculation of mRNA half-lives.
(H) Immunoblot analysis of ribosomes purified by centrifugation through a sucrose cushion with antibodies against GCN1, HSPA8, eRF1, CNOT3, CCT4, RPL29, and RPS5 (left). Blots were quantified by densitometry (right) (n = 3). Error bars represent mean ± SEM. p values by Holm-Sidak’s test.
(I and J) (I) Examples of mRNA turnover profiles of two multipass TMD-encoding transcripts. SLC16A9, 12 TMD segments and (J) SLC9A6, 11 TMD segments. Each time point (0, 2, 4, and 8 h) is represented by dots, and error bars indicate SEM. The lines indicate nonlinear fits and the light-colored shaded area the 95% confidence intervals.
(K) Loss of GCN1 generally dampens codon dependence of mRNA turnover rates, which correlate with codon optimality, represented by a color gradient from blue (optimal) to red (nonoptimal) based on tAI scores. Dotted line indicates no difference in CSC between wild-type and GCN1 knockout cells. Solid line indicates observed change in CSC with shaded area representing the 95% confidence interval.