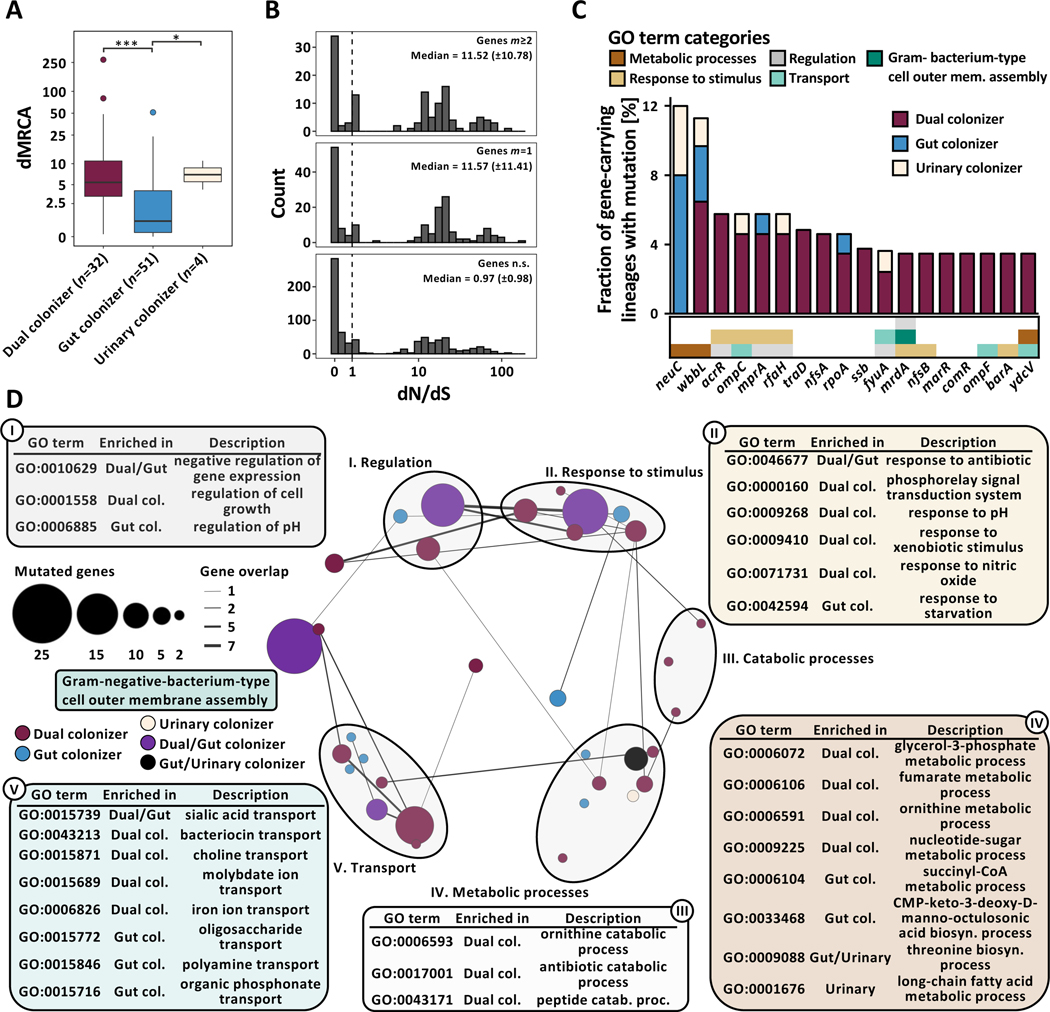
Figure 2 |. Niche-specific adaptation shapes UPEC within-host adaptation.
(A) Boxplot of lineage dMRCA values (n=87 lineages, Kruskal-Wallis P=1.38e−05, Dunn post-hoc test gut vs dual colonizer P=2.39e−05, gut vs urinary colonizer P=3.32e−02). Outliers (outside 1.5x interquartile range) are depicted as points. Whiskers represent 1.5x interquartile range. Upper, middle, and lower box lines indicate 75th, 50th, and 25th percentiles, respectively. (B) Histogram of gene-wise dN/dS values with signatures of non-random mutation (Permutation test, P<0.05) mutated in parallel across more than two lineages (m≥2, top) or in one lineage (m=1, middle), and in genes non-significant in permutation test (bottom). Median and median absolute deviation (MAD) are given for both gene groups. Dashed vertical line indicates neutral selection at dN/dS=1. (C) Genes found to be mutated in parallel in ≥3 lineages, normalized by the total number of gene-carrying lineages. Hypothetical genes are not shown. Color of the bar corresponds to colonization type in which mutations were found (gut colonizer - blue, dual colonizer - maroon, urinary colonizer - light yellow). Color bar below the histogram provides GO category (as shown in Fig 2D) for all genes with GO terms annotation found to be significantly enriched in a colonization type. (D) Network visualization of GO terms significantly overrepresented in the pool of genes with non-random signature of selection within-lineages as defined by the permutation test. Bubble size represents number of mutations in genes categorized into each GO term. Color of bubbles corresponds to colonization type GO terms were enriched in (gut colonizer: blue; dual colonizer: maroon; urinary colonizer: light yellow; gut/dual colonizer: purple; gut/urinary colonizer: black). GO terms were clustered semantically into the 2D space using REVIGO. Circles group together semantically related GO terms.