Figure 5.
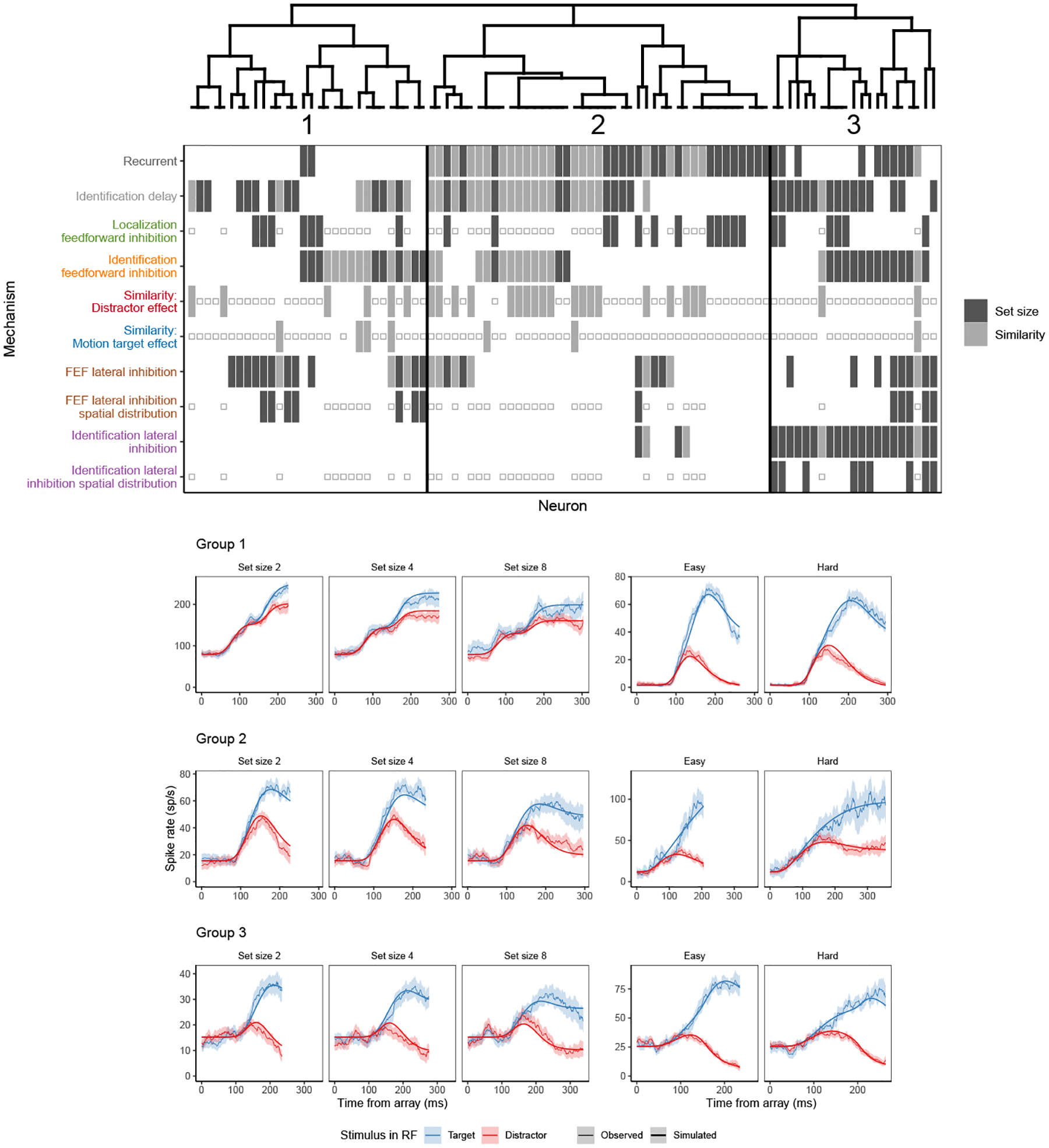
SCRI mechanisms selected by AIC for each neuron. Each column represents the minimal set of mechanisms needed to account for each neuron’s spiking pattern. The presence of a bar indicates that the mechanism was included in the set. If a bar is not present, the parameter corresponding to that mechanism is fixed at zero in the AIC-preferred set. A small open square indicates a mechanism that was not applicable to that neuron, either because it represents an experimental manipulation not performed with that neuron (similarity parameters for neurons recorded under set size manipulations) or because that mechanism was not identifiable given the conditions recorded from that neuron (localization-based feedforward inhibition and spatial distributions for neurons recorded under similarity manipulations). Mechanism labels are colored corresponding to the colors depicting that mechanism in Figure 3. A dendrogram constructed by hierarchical agglomerative clustering based on the AIC-selected mechanisms for each neuron broadly divides neurons into three groups. Below are fits of the full SCRI model to representative neurons (one recorded under set size manipulations, one recorded under similarity manipulations) from each of the three groups. As in Figure 4, for visualization purposes, predicted and observed spike rates were convolved with a kernel representing postsynaptic response (Thompson et al., 1996). Shaded regions depict 95% confidence intervals about the mean.
