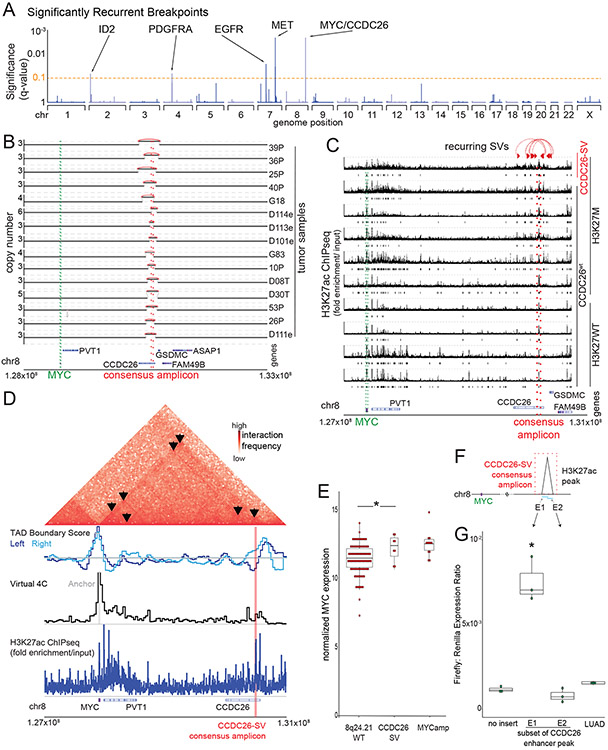
Figure 1. Significantly recurrent breakpoints within CCDC26.
(A) Significance (multiple hypothesis corrected q-values, vertical axis) of recurrent breakpoints (genomic positions on the horizontal axis) across the 179 pHGG genomes. (B) Copy-number profiles across the MYC TAD for the 15 tumors with the recurrent CCDC26 SV. (C) H3K27ac ChIP-seq tracks within the TAD containing MYC (green lines) showing an H3K27ac peak at the location of CCDC26-SV in six H3K27M and four H3K27WT pHGGs tumors. Only the top H3K27ac enrichment track originates from a tumor with a CCDC26-SV. Significantly enriched peaks (q-value < 0.01) are indicated below each H3K27ac ChIP-seq track. The CCDC26 amplicon boundaries for individual samples are indicated by the paired red arrows at the top. The consensus amplicon is indicated by the red dotted lines, and centers on an H3K27ac peak. (D) Hi-C heatmap across the MYC-CCDC26 locus from a midline glioma with the CCDC26-SV. Increasing interaction frequencies are indicated by brighter shades of red. The black arrowheads indicate significant interaction loops. The track beneath the heatmap indicates RobusTAD left and right TAD boundary scores, which represent the likelihood that TAD boundaries are present. The third row contains a virtual 4C track, in which peaks indicate higher interaction frequencies with an anchor sequence in the MYC promoter, which is highlighted in grey. The fourth row shows H3K27ac ChIPseq data from the same sample indicating the location of the enhancer peak within the CCDC26-SV consensus amplicon. (E) Normalized MYC expression in DMG samples with wild-type copy-number profiles at 8q24.21 (n= 92 tumors), CCDC26-SVs (n=8 tumors) or amplifications of the MYC coding sequence (n=12 tumors). *denotes p = 0.04 as determined by two-sided Wilcoxon rank sum test. Center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5x inter-quartile range (IQR). (F) Schematic illustrating the luciferase reporter used to validate the enhancer in CCDC26, showing the positions of the E1 and E2 sequences with respect to the enhancer within CCDC26. (G) Luciferase activity in DIPG13 cells following transduction of the E1, E2, and LUAD enhancer reporters or empty vector controls. Values represent the average of four technical replicates in each of three independent experiments. *denotes p = 1.6x10−5 (E1 vs Backbone) and p = 0.89 (LUAD vs Backbone), n = 3 independent experiments, Nested One-Way Anova with Tukey’s post-test, boxplot defined as in E.