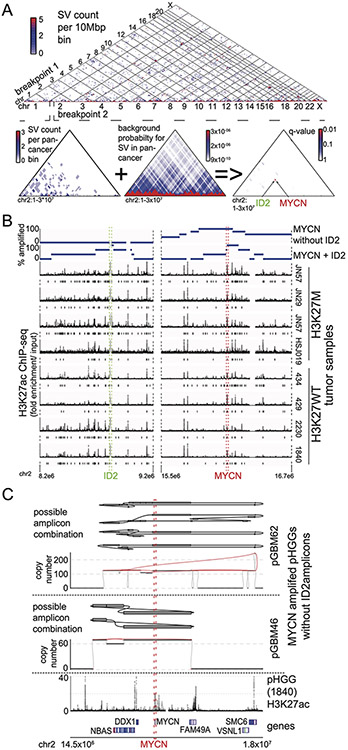
Extended Data Fig. 3. Significantly recurrent juxtaposition between MYCN and ID2.
(A) (top) Count matrix showing all possible juxtapositions between pairs of genomic loci. (bottom) Illustration of the principle behind the analysis of recurrent juxtapositions, as exemplified by the MYCN-ID2 loci. First, we count the number of SVs connecting each pair of genomic loci. Using a background model for the probability of juxtapositions generated from an analysis of 2658 cancers21, we then determine the probability of observing this number of SVs due to chance alone, corrected for multiple hypothesis testing. This analysis revealed the MYCN-ID2 juxtaposition as the only significantly recurrent juxtaposition in the window shown. (B) Overlay of amplification frequencies on ChIP-seq data in the ID2 and MYCN loci. The top two tracks show, among tumors with MYCN-ID2 rearrangements (top track, n=4 tumors or MYCN amplifications without ID2 involvement (second track, n=4 tumors), the percentage of tumors with amplifications (y-axis) at each genomic locus (x-axis). The bottom eight tracks indicate H3K27ac ChIP-seq profiles across these loci for four H3K27M and four H3WT pHGGs tumors. Coding sequences of ID2 and MYCN are highlighted with yellow and red lines respectively. Significantly enriched H3K27ac peaks (q-value < 0.01) are indicated below each ChIP-seq track. The small region at the ID2 locus that is amplified in all MYCN-ID2 pHGGs shows an H3K27ac peak in the ChIP tracks from all six pHGG tumor samples. Tumors that amplify MYCN without ID2 take in a much larger region of the MYCN TAD into the amplicon. (C) G-track plots indicating copy-number profiles and genome topology after consideration of local SVs, for two examples of pHGGs with focal MYCN amplicons without incorporation of ID2. For both tumors the copy number and SV profiles support several possible reconstructions of extrachromosomal circular amplicons. All are limited to the neighborhood of MYCN, presumably incorporating endogenous enhancers from the MYCN TAD.