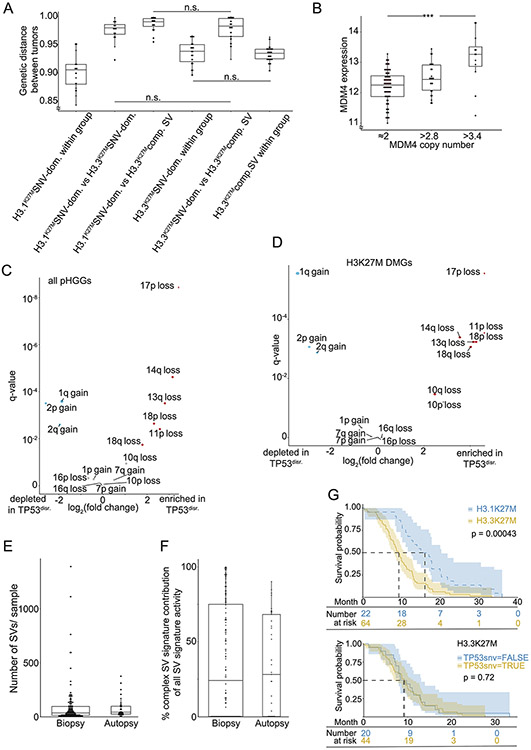
Extended Data Fig. 9. Associations between SV + histone defined groups.
(A) Jaccard distances between tumor pairs (vertical axis), calculated from the combination of variants in each tumor, across subgroups of H3K27M DMGs. Tumor groups were determined by H3.1 or H3.3 mutations and the combined complex SV signatures exceeding 20% of all SV signature activity. Tumors were paired within or between these groups, as indicated on the horizontal axis. All differences were significant with q < 0.005 (FDR-corrected two-sided Wilcoxon test) unless indicated otherwise. n = 165 DMGs) (B) Association between MDM4 expression (vertical axis) and copy number (horizontal axis). MDM4 gains universally represent arm-level gains of 1q. *** indicates adjusted p = 0.004, ANOVA with two-sided Tuckey post-test, n = 114 tumors. (C-D) Volcano plot indicating the significance (vertical axis; FDR-corrected Fisher’s exact tests) of associations between genetic variants and pHGG subgroups (horizontal axis). (C) Arm-level SCNAs in TP53-disrupted (n=97 tumors) vs TP53WT pHGGs (n = 77 tumors). TP53 disruption represented SNVs (n=88 tumors, often with copy loss) or copy loss alone (n=9 tumors). (D) Arm-level SCNAs in TP53-disrupted (n=56 tumors) vs TP53WT H3K27M mutant DMGs (n = 39 tumors). TP53 disruption represented SNVs (n=53 tumors often with copy loss) or copy loss alone in (n=3 tumors). Only significantly recurrent arm-level SCNAs are shown. (E-F) Comparison between pre-treatment biopsy and autopsy samples. These groups exhibit no significant differences in (E) the number of SVs per sample (q= 0.6, Wilcoxon, n= 174 tumors) or (F) the activity of the combined complex-SV signatures (q= 0.7, Wilcoxon, n= 174 tumors). Center line of the boxplots indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR. (G) Kaplan-Maier plot indicating overall survival for (top) H3.1K27M and H3.3K27M DMGs and (bottom) H3.3K27M DMG with and without TP53 SNVs. p-values are from log-rank tests. Error bands show the 95% confidence interval.