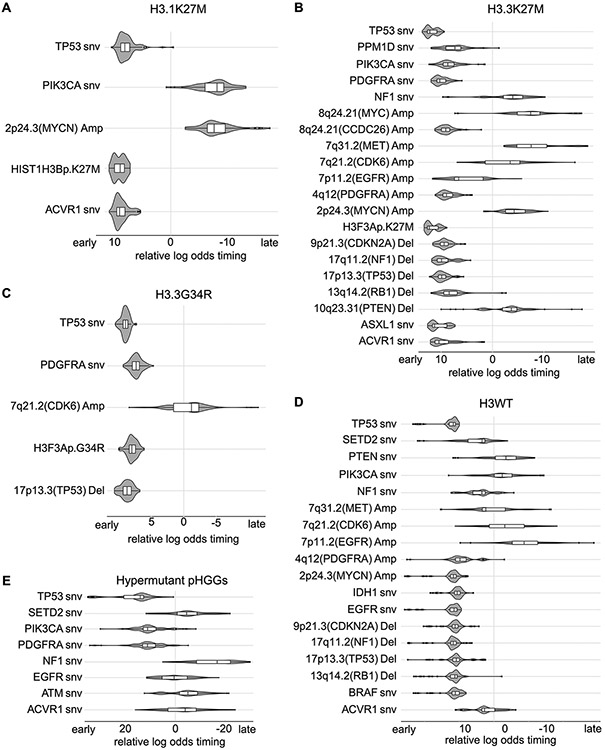
Extended Data Fig. 10. Timing analysis of somatic variant acquisition in histone mutation-defined pHGG subgroups.
For each subgroup the individual (per-sample) timing of recurrent variants is fed into a Bradley-Terry model. This results in a strength parameter for each variant which is indicated on the x-axis in log scale and can be interpreted as the relative log odds of the variant being an early event in this subgroup. Each distribution indicates the results of 100 random subsamples of the respective subgroup. Only potential driver variants recurrent in more than two samples are shown. Subgroups: (A) H3.1K27M, n = 24 tumors (B) H3.3K27M, n = 73 tumors (C) H3.3G34R, n = 14 tumors (D) H3WT, n = 63 tumors (E) hypermutant pHGGs, n = 5 tumors. Center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR