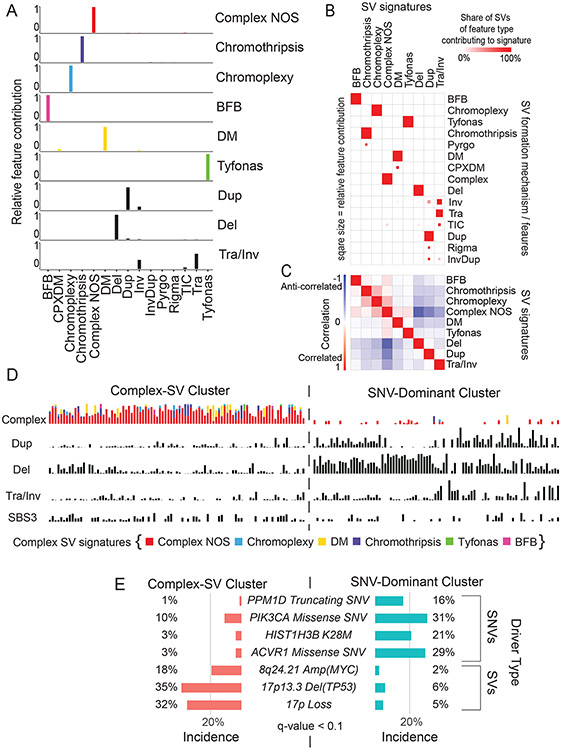
Figure 4. SV signatures in pHGG.
(A) Contribution of different SV features to each of the identified SV signatures. The horizontal axis indicates SV features - Deletions (Del), duplication (Dup), inversion (Inv), breakage fusion bridge cycle (BFB), complex double minute (cpxdm), double minute (dm), template insertion chains (tic), and translocation (tra). The vertical axis indicates the fraction of SVs with each of these features within the nine identified SV signatures. (B) Heatmap indicating the contribution of each SV type to each signature. Deeper red color indicates a larger fraction of all SVs with each type contributing to each signature. The size of the squares indicates the fraction of all SVs contributing to a signature that belong to this SV type. (C) Heatmap indicating the correlation between the SV-signatures as determined by Pearson. Shading in red indicates positive correlation coefficient and blue indicates anti-correlation. (D) Normalized SNV and SV signature activities within the Complex-SV and SNV-Dominant clusters. Each column represents an individual tumor and rows indicate signature activities in the individual samples. Contribution of complex SV signatures within the Complex SV group are represented by the colors shown (n=179 tumors). (E) Genetic variants (SV and SNVs) significantly enriched in the Complex-SV and SNV-Dominant clusters (n=179 tumors), with correlations reaching q < 0.1. q-values were calculated using Fisher’s exact tests.