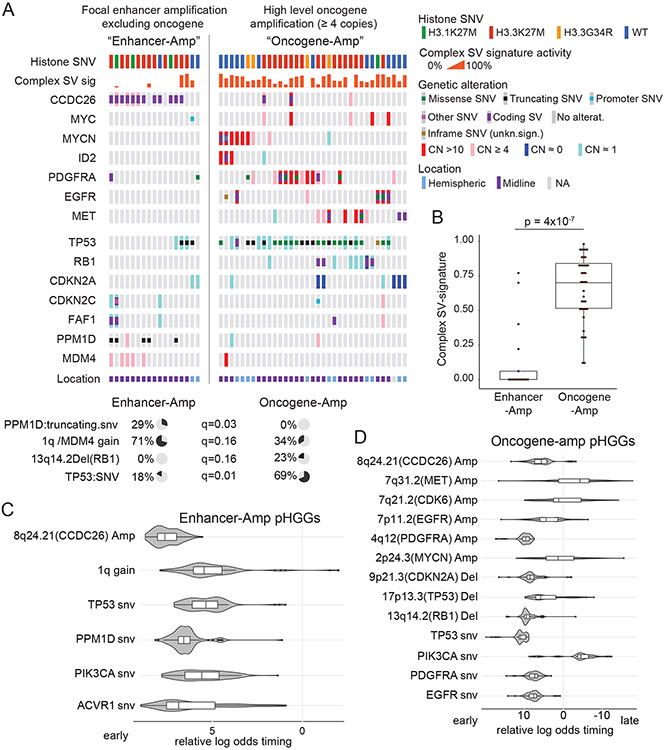
Figure 6. Context of the significantly recurrent SVs.
(A) Co-mut plot for Enhancer-Amp and Oncogene-Amp significantly recurrent SVs. Enhancer-Amp significantly recurrent SVs generate focal amplifications in the TAD of an oncogene without amplifying the protein coding sequence; Oncogene-Amp significantly recurrent SVs generate high-level (CN >3.4) amplifications or fusions of the coding sequence. The top two rows show associated metadata. The next seven rows indicate the genes affected by the significantly recurrent SVs. The bottom seven rows show genes in DNA damage response pathways. Significant associations with the two groups are illustrated with pie charts below the plot, based on Fisher’s exact tests. (B) Enhancer-Amp pHGGs show significantly lower combined Complex-SV signature activity than Oncogene-Amp pHGGs (p = 4x10−7, two-sided Wilcoxon, n = 52 tumors, center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR). (C-D) Timing analysis of somatic variant acquisition in Enhancer-Amp (C) and Oncogene-Amp (D) pHGGs based on a Bradley-Terry model. The horizontal axis shows the log odds of the variant being an early event. The distributions indicate the results of 100 random subsamples of the data (C: n = 17 pHGGs, D: n = 35 pHGGs). Center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR. Only variants in the SRSV-affected genes and pathways (growth factor and MYC signaling) and in DNA damage response genes altered in more than two samples are shown.