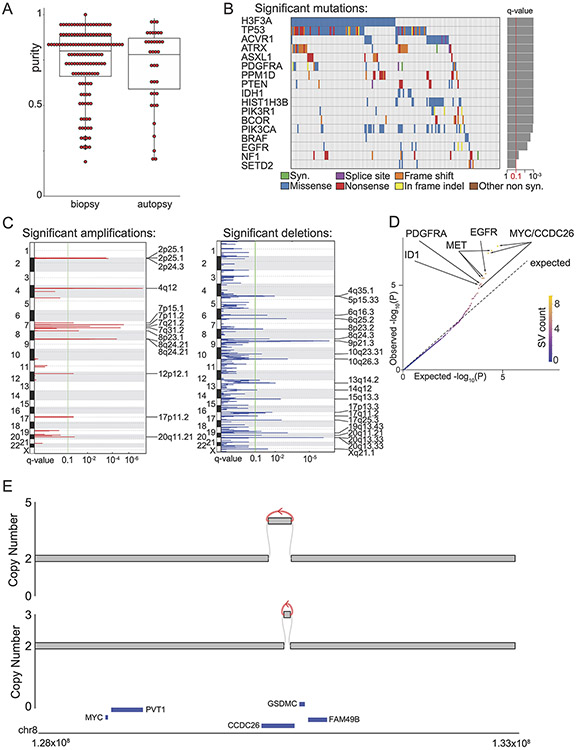
Extended Data Fig. 1. Sample characteristics and significantly recurrent variants.
(A) Purity of pre-treatment biopsy and autopsy samples were not significantly different (p = 0.5, two-sided Wilcoxon, n= 174 tumors, center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR). (B) Significantly recurrent SNVs in non-hypermutant tumors (n= 179 tumors). (C) Significantly recurrent SCNAs (n= 179 tumors). All of these SCNAs have been noted3 except for a non-protein-coding locus in 8q.24.21, near MYC—which is also within a separate recurrently amplified locus. (D) Q-Q plot for the analysis of significantly recurrent SV breakpoints. The most significantly recurrent breakpoints are within the long non-coding RNA CCDC26, within the TAD encompassing MYC (based on n= 179 tumors). (E) Representative examples of the enhancer amplification through simple tandem-duplications within the long non-coding RNA encoding CCDC26.