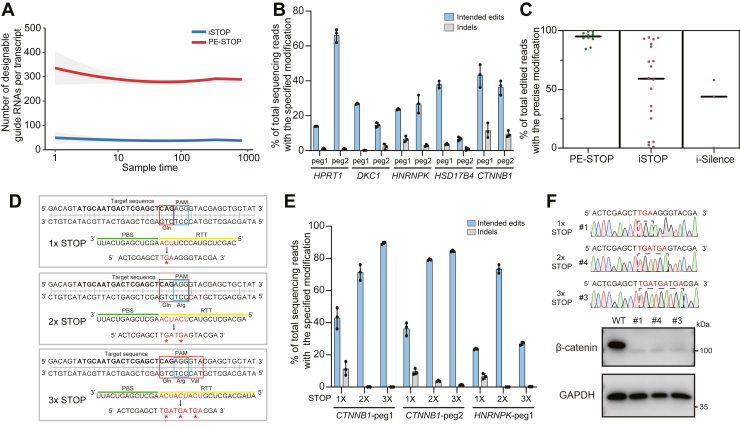
Figure 3.
PE-STOP allows efficient nonsense substitutions within genomic contexts.A, the graph shows the predicted numbers of stop codons programmable by pegRNAs (PE-STOP, red) or sgRNAs (iSTOP, blue) on an average human transcript. A given number of transcripts were randomly sampled for three times and were used to model the overall numbers of editable sites via locally estimated scatterplot smoothing. B, the editing efficiencies by PE-STOP for targeting five genes (two sites/gene) are shown. The editing efficiencies and the associated indel levels were determined from the deep sequencing data (n = 3 biological replicates). Data are presented as mean values ± SD. C, the on-target editing purities by PE-STOP, iSTOP, and i-Silence are presented. The precise editing efficiencies are indicated by the percentages of reads harboring only the targeted mutations (perfect edits) within all edited reads. Each data point shown in the scattered plot represents the averaged value (from three biological replicates) for a single site. The horizontal lines in each editing group represent the median levels of precisions for each editing tool. D, as an example, the design of PE-STOP for installing single (1x), double (2x), or triple (3x) stop codons in CTNNB1 (peg1 site) is illustrated by the layouts of target and pegRNA sequences. The PAM motifs are highlighted by the blue boxes, whereas the targeted codons are highlighted by the red boxes. The PBS (green line) and RTT (yellow line) sequences within respective pegRNAs for installing 1x, 2x, and 3x stop codons are shown, with the edits marked in red. E, the efficiencies and the associated indel levels by PE-STOP for installing single (1x), double (2x), or triple (3x) stop codons at three different sites (within two genes) are shown. For installing various numbers of stop codons, the pegRNAs featured the same spacer and PBS sequences but different RTT sequences encoding the edits. Data were obtained from three biological replicates (shown as the mean ± SD). F, HEK293T cells were transfected with PE-STOP plasmids to program single, double, or triple stop codons in the gene encoding β-catenin (CTNNB1, “peg1” site as in [D and E]). Following transfection, single clones were cultivated for subsequent determination of genotypes. The Sanger sequencing results from clones corresponding to the homozygous mutants with various forms of nonsense mutations (1× STOP, 2× STOP, and 3× STOP) are presented on the top. The levels of protein expression for β-catenin in cells derived from the WT and mutant clones were determined by Western blot, with the use of GAPDH as loading control (lower). HEK293T, human embryonic kidney 293T cell line; PBS, primer binding site; pegRNA, prime editing guide RNA; RTT, reverse transcription template; sgRNA, single guide RNA.