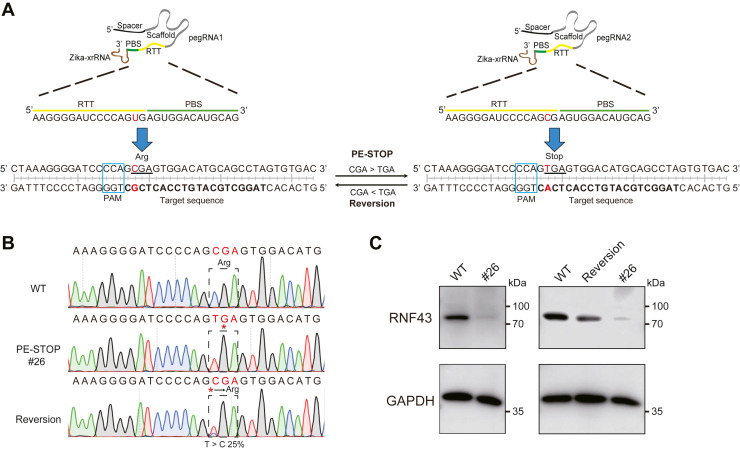
Figure 4.
PE-STOP-induced nonsense mutations may be subjected to subsequent precise reversions by the same PE platform.A, the design of PE for successive nonsense and reverse mutation is illustrated by the layouts of target sequences (in RNF43) and the corresponding pegRNAs. The sequences from the WT and the nonsense mutant allele (R519X) are placed on the left and right sides, respectively. The pegRNA1 is designed to install the nonsense mutation, whereas the pegRNA2 is adopted to enable the reverse mutation. The RTT (yellow line) and PBS (green line) sequences within respective pegRNAs are shown. On the WT and mutated (R519X) sequences, the PAM motifs are highlighted by the blue boxes, and the targeted codons are underlined. B, HEK293T cells were first transfected with PE-STOP (in PE3 format) to install R519X mutation. Following sorting the transfected cells into single clones, a clone (#26) with homozygous nonsense mutation was identified by Sanger sequencing (middle panel). The cells derived from this clone were transfected again with PE to drive the reverse mutation. The transfected cells were pooled by fluorescent marker sorting. This mixed population of cells were further expanded in culture for 7 days. The DNA samples were harvested. Visible levels of reverse mutation can be captured by Sanger sequencing (bottom panel). C, the protein samples were harvested from cells derived from the WT clone and the nonsense mutant clone (#26). Following transfection of the mutant cells with the back-mutation PE plasmids for 48 h, the transfectants were harvested via FACS. The cells were further expanded for 7 days. The total protein samples were harvested. The indicated protein samples were analyzed by WB. FACS, fluorescence-activated cell sorting; HEK293T, human embryonic kidney 293T cell line; PBS, primer binding site; PE, prime editor; pegRNA, prime editing guide RNA; RTT, reverse transcription template; WB, Western blot.