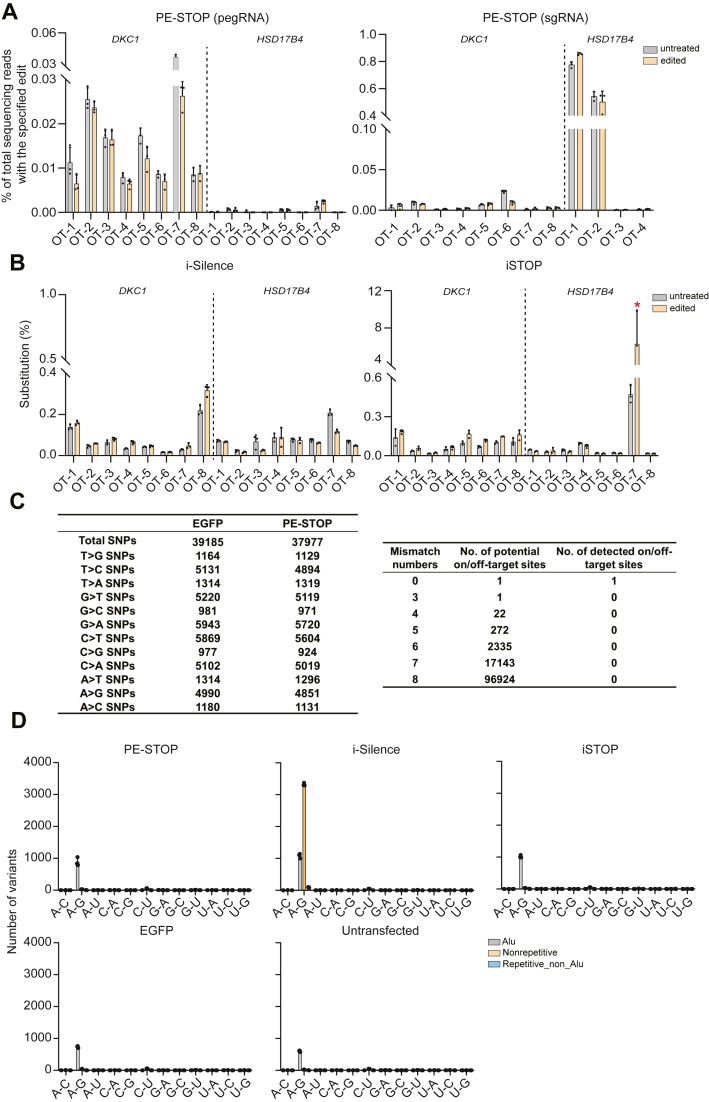
Figure 5.
PE-STOP exhibited better specificity profiles than iSTOP and i-Silence.A, DNA off-target (OT) analyses were performed following the application of PE-STOP against DKC1 (peg1) and HSD17B4 (peg1). Targeted deep sequencing experiments were performed for the pegRNA (left)- and nick sgRNA (right)-associated potential OT sites. The OT sites analyzed here were a collection of high-score sites predicted by the Cas -OFFinder program. Samples from untreated cells were used as controls. The levels of sequence variances at these sites are presented in graphs (with indications of their corresponded pegRNAs [target genes]). The average values from three biological replicates are shown (±SD). B, DNA OT analyses were performed following the application of i-Silence (left) and iSTOP (right) against DKC1 (sgRNA1) and HSD17B4 (sgRNA1). Targeted deep sequencing experiments were performed for the sgRNA-associated potential OT sites. Samples from untreated cells were used as controls. The levels of base substitutions corresponding to ABE or CBE activities at these sites are presented in graphs (with indications of their corresponded pegRNAs [target genes]). The average values from three biological replicates are shown (±SD). An apparent indication of iSTOP-dependent OT editing (C-to-T) is highlighted by a red mark. C, WGS analyses were performed following the application of PE-STOP against HSD17B4 (peg1). The EGFP-transfected group was used for comparisons. The reads from untransfected cells were used to filter out the basal genetic variances in both groups. In addition, sites with 0 to 8 mismatches from the targeted site were queried for edits. On the left, the numbers of de novo SNPs in these two groups are displayed for indicated substitution types. The results on the right show identification of only the desired edit by WGS. D, potential editing by different editors on cellular RNA were assessed by analyses of RNA-Seq data. Numbers of all RNA sequence variants with base substitutions were determined in samples from cells introduced with different editors (PE-STOP [PEmax + pegRNA + nick sgRNA], iSTOP [AncBE4max + sgRNA], and i-Silence [ABE8e + sgRNA]). Samples from the untransfected and EGFP-transfected cells were also used for comparisons. The numbers of variants are grouped in accordance with indicated repetitive or nonrepetitive features. The average values from three biological replicates are shown (±SD). ABE, adenine base editor; CBE, cytosine base editor; EGFP, enhanced GFP; pegRNA, prime editing guide RNA; sgRNA, single guide RNA; WGS, whole-genome sequencing.