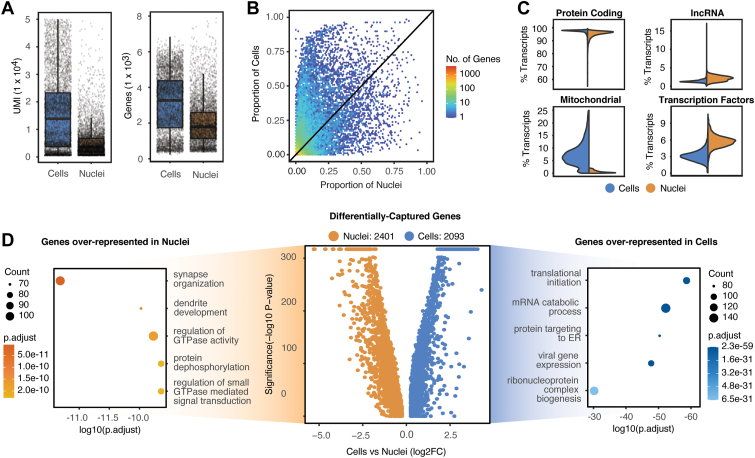
Figure 3.
Differences between single-cell and single-nucleus RNA-sequencing modalities. A, Boxplots showing that the unique molecular identifier (UMI) capture rate (left) and the gene capture rate (right) is higher in cells versus nuclei. B, Binned scatterplot of the proportion of genes detected in cells vs. nuclei. C, Split violin plots displaying the distribution of percentage transcripts that map to protein coding (top left), long noncoding RNA (top right), mitochondrial (bottom left), and transcription factor genes (bottom right). D, Four thousand four hundred ninety-four genes are differentially captured in the combined data set with 2401 genes expressed higher in nuclei and 2093 expressed higher in cells. The top 5 overrepresented gene ontology terms (GO) are displayed on the left (nuclei) and right (cells) of the volcano plot.