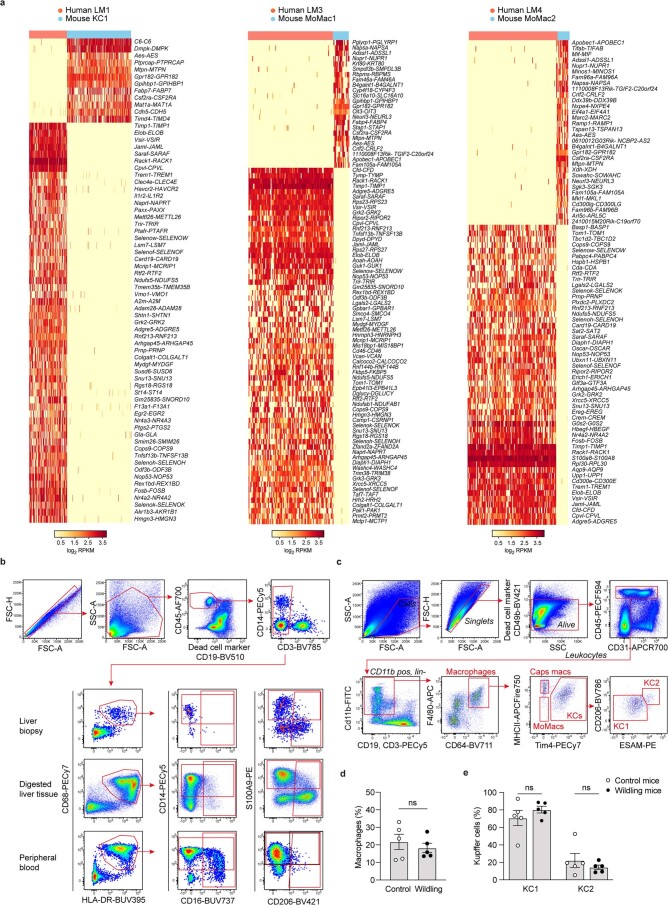
Extended Data Fig. 6. Human-mouse conservation of the LM cell populations.
a, Heatmaps of genes specifically expressed (log2 RPKM) by humans or by mice for each matched LM cell subpopulation present in both mice and humans. b, Gating scheme for flow cytometry analysis of human livers and intrahepatic blood, where cells were gated amongst all live CD45+CD68+HLA-DR+ myeloid cells. LM1 was defined as CD14+CD16+CD206+S100A9-, LM2 as CD14+CD16-CD206+S100A9-, LM3 as CD14+CD16+CD206-S100A9+, and LM4 as CD14+CD16-CD206-S100A9+. c, Gating scheme for flow cytometry analysis of macrophage subpopulations in murine livers of wildling and specific pathogen-free (SPF) control mice, gated using FMO controls. d, Proportion of murine liver macrophages in wildling (n=5) and SPF control (n=5) livers. Liver macrophages are gated as live CD45+ lineage-(CD49b, CD3, CD19) CD11b+ F4/80+ CD64+. e, Proportion of murine KC subsets in wildling (n=5) and SPF control (n=5) livers. KC1 is gated as live CD45+ lineage-(CD49b, CD3, CD19) CD11b+ F4/80+ CD64+ Tim4+ CD206- ESAM- and KC2 is gated as CD45+ lineage-(CD49b, CD3, CD19) CD11b+ F4/80+ CD64+ Tim4+ CD206+ ESAM+. Data are presented as mean ± s.e.m. P values were calculated by two-tailed Mann-Whitney test (d) or two-way ANOVA with adjustment for multiple comparisons (e). *p < 0.05. RPKM, Reads per kilobase of exon model per million mapped reads; LM, liver myeloid cells; KC, Kupffer cell; MoMacs, monocyte-derived macrophages; Caps macs, capsular macrophages.