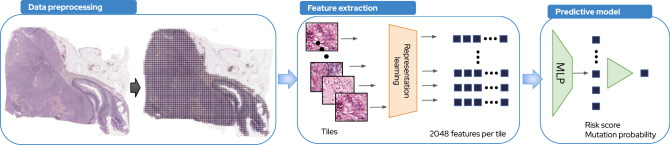
Fig. 1. Deep learning workflow.
Our DL pipeline consists of three main steps. Data preprocessing. For one raw WSI, we apply a matter detection algorithm on it in order to extract the tissue area and to remove the artifacts (blur area, etc). On the extracted tissue area, we apply a tiling which consists of dividing the whole-slide images into tiles of 112 × 112 μm (224 × 224 pixels) at a zoom level of 0.5 microns per pixel. Feature extraction. We trained from scratch a 50-layer ResNet using an inhouse dataset of sarcoma of 1287 WSI (942,626 tiles) and Momentum Contrast v2 (MoCo v2) algorithm, a self-supervised learning algorithm based on contrastive loss. Using this model, we extracted 2048 features from each of the tiles, such that a slide could be represented as a N × 2048 matrix with N equals to the total number of tiles. Predictive models. MLP were trained using the 2048 features to predict mutation types or survival risk.