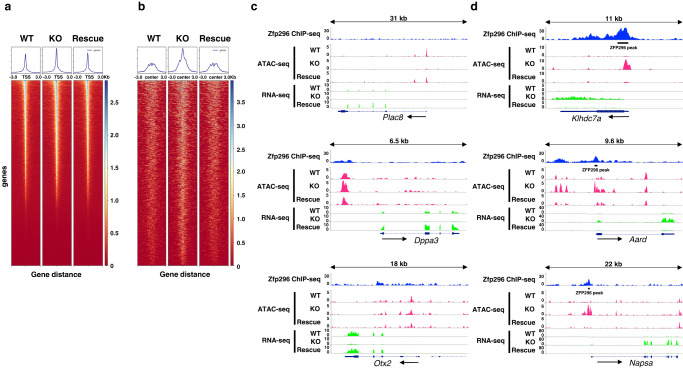
Fig. 5. Analysis of chromatin accessibility around TSS and ZFP296 binding sites.
a Heatmaps and line plots showing average ATAC-seq signal centered on TSS. Each row corresponds to an individual gene. b Heatmaps and line plots showing average ATAC-seq signal centered on ZFP296 binding peaks located within 3 kb from TSS. Each row corresponds to an individual binding site. Heatmaps and line plots were made using deepTools. c, d Genome browser tracks of counts per million (CPM) mapped reads of ZFP296 ChIP-seq and ATAC- and RNA-seq (WT, KO #98, and Rescue #22) across three genes each that showed a marked decrease and increase in expression after Zfp296 KO.