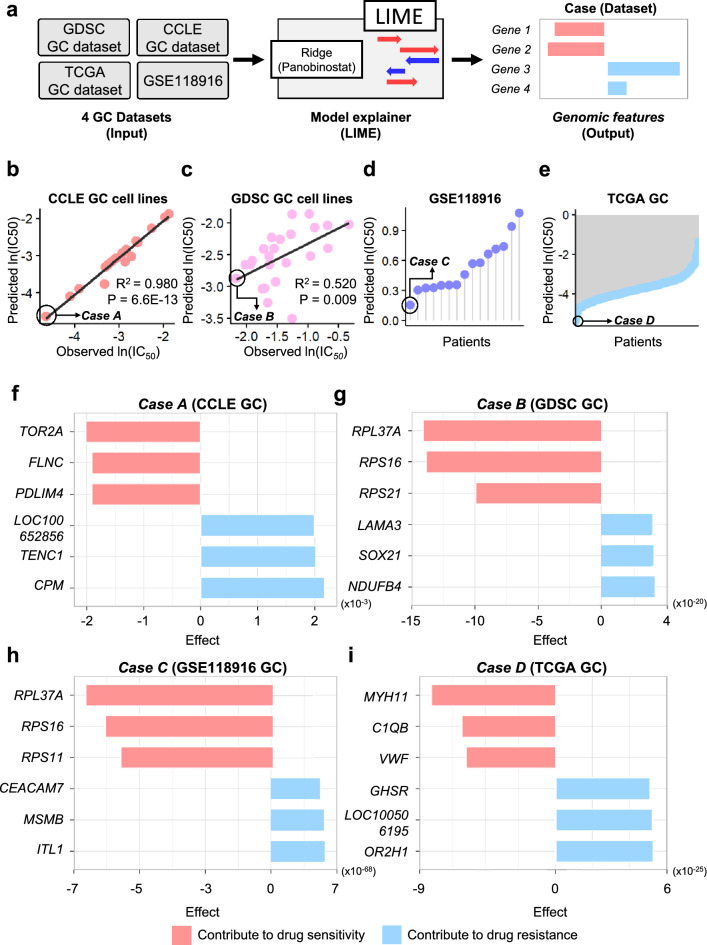
Figure 4.
Application of the ridge model for panobinostat in setting 1 to the GC cell line (CCLE and GDSC) and patient datasets (GSE118916 and TCGA). (a) Description of XAI application. (b) CCLE and (c) GDSC GC cell lines show a good correlation between predicted and observed ln(IC50) values. (d) GSE118916 and (e) TCGA datasets for patients with GC show a broad distribution of predicted ln(IC50) values. Furthermore, we selected a sensitive case for panobinostat in each GC dataset, and cases A through D are indicated in the circle. For case A and B, the predicted IC50 values of the two cases agree with their observed IC50 values. Considering case C and D, their datasets did not have patient information on the response, and the prediction cannot be compared. Considering prediction, the four cases are sensitive to the panobinostat when compared with the other samples. (f–i) To identify major genes affecting drug response, we performed XAI analysis with the four cases (case A through D). The six genes affecting drug sensitivity (red bar) and drug resistance (blue bar) were detected in (f) case A, (g) case B, (h) case C, and (i) case D, respectively. CCLE Cancer Cell Line Encyclopedia, GDSC genomics of drug sensitivity in cancer, GC gastric cancer, TCGA The Cancer Genome Atlas, XAI explainable artificial intelligence.