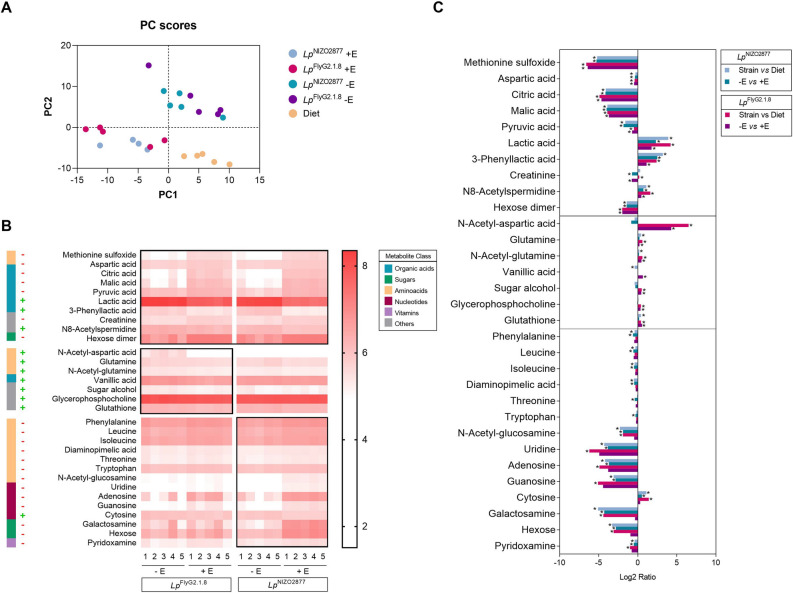
Figure 4.
Metabolic differences between active and stable bacteria. (A) Principal Component Analysis (PCA) of the 5 experimental conditions based on the metabolic profiles. As expected, the presence of antibiotic discriminates samples on both axes (PC1 and PC2). (B) Heatmap showing the metabolites that differ significantly between experimental groups (− E—without erythromycin–and + E—with erythromycin) for each strain tested (LpFlyG2_1.8 and LpNIZO2877) (two-sided t tests, p < 0.05). The compounds are ordered by the metabolite class given by the left scale. Metabolites whose pattern is conserved between strains are framed together in the upper group. The scale on the right indicates the gradient of metabolite presence (i.e., relative concentration of each metabolite detected by LC–MS method) (C) Mean ratio fold-change (plotted on a Log2 scale) of metabolites with statistically significant differences (y axis) between experimental conditions (see legend; *p < 0,001). Fold changes and p-values are provided in Supplementary Table 3.