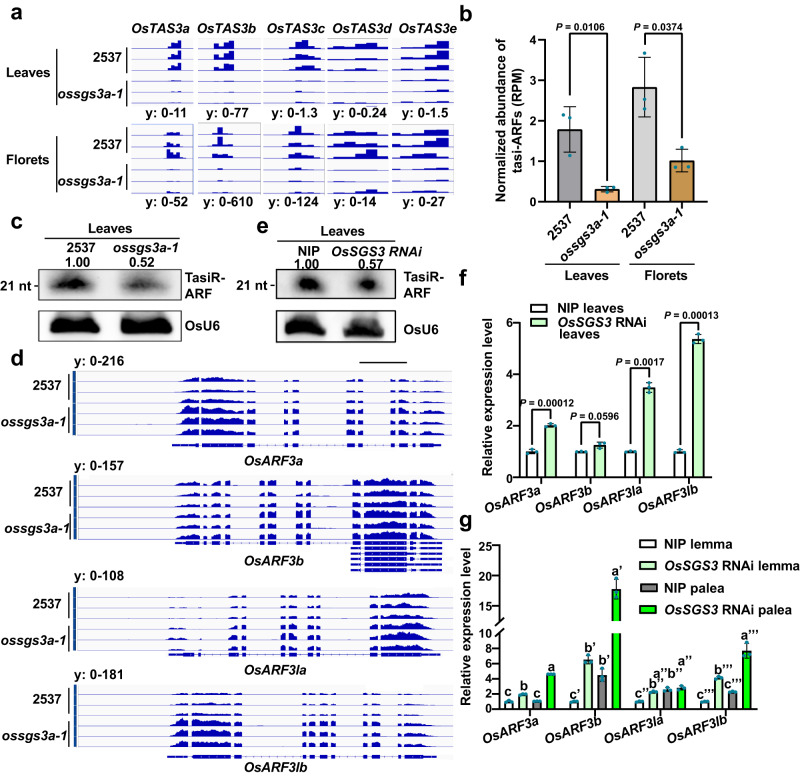
Fig. 3. OsSGS3a modulates the biogenesis of tasiR-ARF.
a Genome browser view of OsTAS3a-e-derived tasiRNAs in the leaves and florets of 2537 and ossgs3a-1. Leaves were sampled from 2-week-old seedlings grown in the growth chamber at 28 °C and florets were collected from 2537 and ossgs3a-1 grown under high field temperature. b Normalized abundance of tasiR-ARFs in the leaves and florets of 2537 and ossgs3a-1 as determined by small RNA deep sequencing. c RNA blot of tasiR-ARFs in the leaves of 2537 and ossgs3a-1 grown in the field. d Genome browser view of the OsARF3la, OsARF3lb, OsARF3a, and OsARF3b mRNAs in the leaves of 2537 and ossgs3a-1. Scale bar, 1 kb. e The abundances of tasiR-ARFs in the leaves of NIP and OsSGS3 RNAi transgenic plants grown in the field. OsU6 was used as a loading control (c, e). The intensity of the blots was quantified (c, e). f, g Real-time RT-PCR analyses of relative expressions of OsARF3la, OsARF3lb, OsARF3a, and OsARF3b in the indicated samples grown in the field. OsActin served as an internal control (f, g). The average values (±s.d., n = 3, biologically independent samples) are shown (b, f, g). Significant differences were determined by two-tailed Student’s t test (b, f) or one-way ANOVA with Tukey’s HSD post hoc analysis (g). Exact P values are indicated above the bars (b, f) or provided in Supplementary Data 7 (g). Experiments were repeated three times with similar results (c, e–g). Source data are provided as a Source Data file (b, c, e–g).