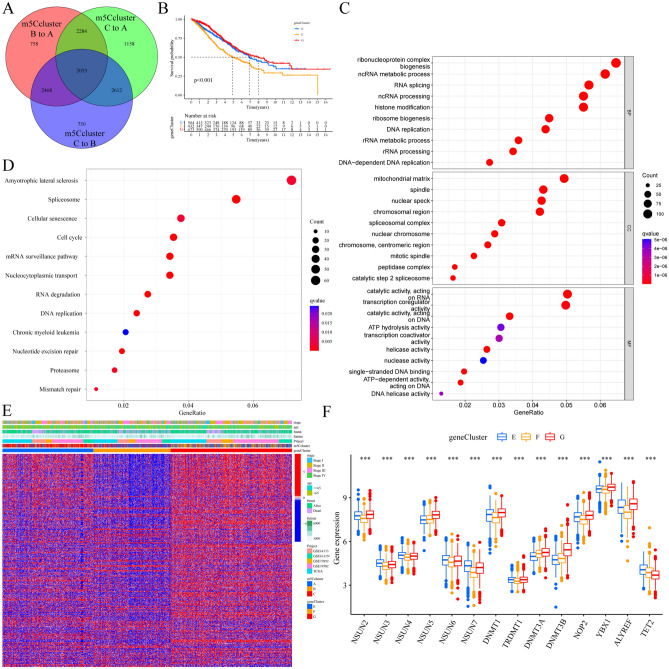
Figure 4.
Construction of an m5C modification phenotype-related differentially expressed gene (DEG) signature and functional annotation in the meta-CRC cohort. (A) A total of 2053 DEGs between the three m5C clusters were identified and are shown in the Venn diagram. (B) The survival curves of the distinct m5C gene signature groups were analyzed by the K-M method (log-rank test). The numbers of patients with the gene cluster E, gene cluster F and gene cluster G phenotypes were 504, 432 and 673, respectively. (C) GO enrichment analysis and (D) KEGG enrichment analysis were performed; functional annotation for m5C modification phenotype-related DEGs is shown (www.kegg.jp/kegg/kegg1.html). The size of the circles represents the enrichment of the genes, and the color depth represents the q-value. (E) Unsupervised clustering of the m5C-related genes into clusters in the five independent CRC cohorts was performed. The clinical characteristics of CRC cohort patients in distinct gene clusters are shown. Each column represents a patient, and each row represent a DEG. (F) Differential expression of 14 m5C regulators between distinct gene cluster groups (*P < 0.05; **P < 0.01; ***P < 0.001).