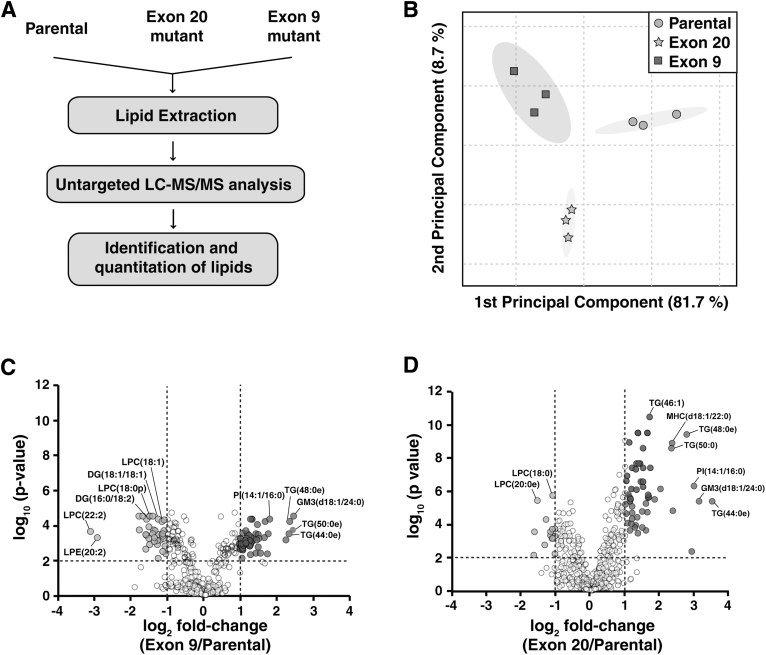
FIG. 1.
(A) Experimental scheme of the untargeted lipidomics workflow. (B) Principal component analysis based on profiling of lipids between parental (circle), Exon 9 (square), and Exon 20 (triangle) cells. (C) Volcano plots of lipid species detected from Exon 9 versus MCF10A and (D) Exon 20 versus MCF10A. Circles in dark gray indicate species that were significantly increased (adjusted p < 0.01 and fold-change >2), and circles in light gray indicate species that were significantly decreased (adjusted p < 0.01 and fold-change <0.5) in Exon 9 or Exon 20 compared with parental MCF10A cells. Circles in white indicate species that remained statistically unchanged in mutants. DG, diglyceride; GM3, monosialogangliosides; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; MHC, monohexosylceramide; PI, phosphatidylinositol; TG, triglyceride.