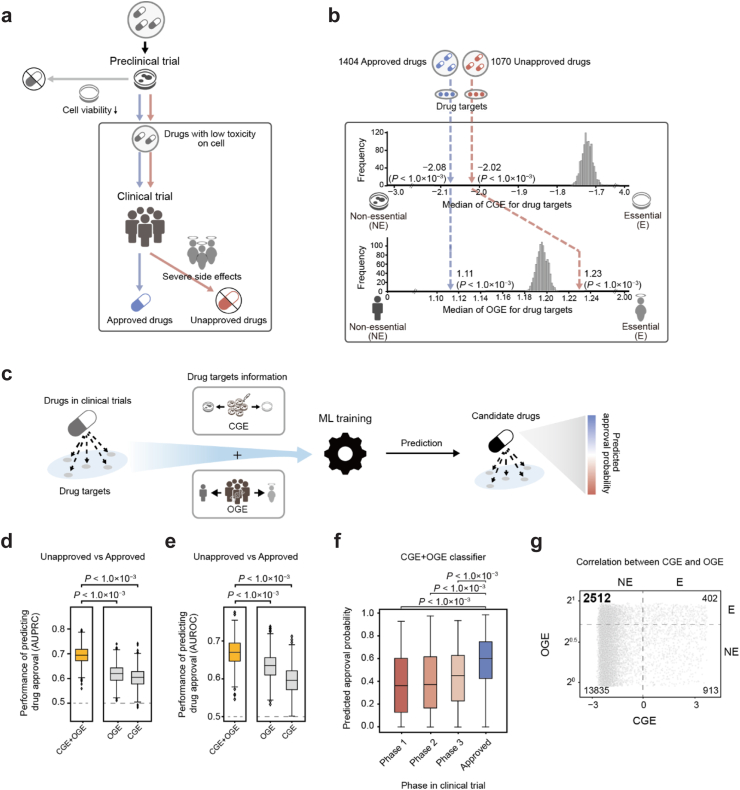
Fig. 1.
Predicting drug approval in clinical trials with the CGE and OGE. (a) Schematic for the drug development process. Drugs in clinical trials passed cell viability tests in preclinical trials. Drugs that failed clinical trials due to severe side effects are shown as unapproved drugs. Drugs that passed clinical trials without severe side effects are shown as approved drugs. (b) The CGE and OGE were mapped onto the unapproved and approved drugs. The numbers of unapproved and approved drugs are shown. The actual median CGE and OGE of 1070 unapproved and 1404 approved drugs are depicted with red and blue dashed lines, respectively. The histograms show the random distributions of median CGEs and OGEs for drugs in clinical trials (random shuffling of CGE and OGE 1000 times). Statistical significance (P) was evaluated using a two-tailed test. (c) The overall framework for predicting drug approval using the CGE and OGE of drug targets with machine learning (ML). (d–e) The AUPRC and AUROC of the CGE + OGE, OGE, and CGE classifiers for predicting drug approvals in clinical trials. Unapproved drugs (drugs with max phase 1, 2, and 3) and approved drugs were predicted. Box plots represent the AUPRC and AUROC for Monte Carlo cross-validation (1000 times). Statistical significance (P) of the differences between AUPRCs/AUROCs was analysed using the Mann–Whitney U test. The grey dashed line denotes the baseline. (f) Distribution of predicted approval probabilities according to the clinical trial phase. Statistical significance (P) of the difference in predicted approval probabilities between clinical phases was analysed using the Mann–Whitney U test. (g) The numbers of genes in each area are indicated. Dashed lines denote the thresholds of the CGE and OGE to discriminate between essential and non-essential genes. The y-axis was set to the log2 scale. CGE, cellular gene essentiality; OGE, organismal gene essentiality.