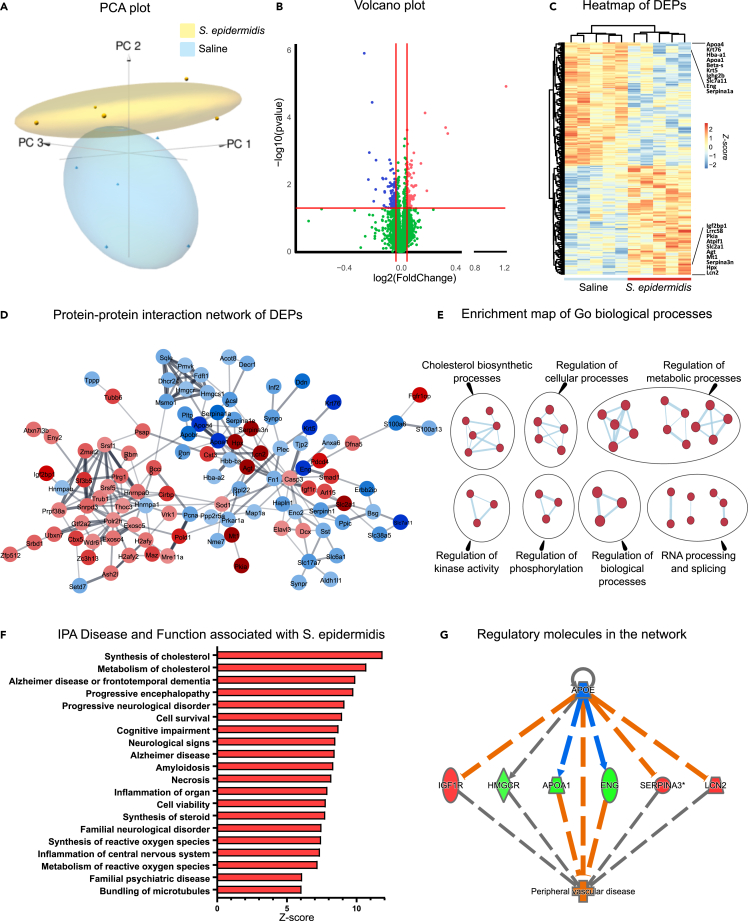
Figure 1.
Changes in hippocampal protein expression following S. epidermidis infection
(A) Principal component analysis of hippocampal proteomics shows a clear separation of samples from animals treated with saline and S. epidermidis at 24 h post-infection (saline n = 5, S. epidermidis n = 5).
(B) Volcano plot showing the spread of 5914 differentially expressed proteins (71 downregulated, blue and 78 upregulated, red, see also Table S1).
(C) Heatmap of differentially expressed proteins. The top 10 down- and up-regulated proteins are enlarged and listed in the heatmap.
(D) Protein-protein interaction network constructed from the 149 DEPs. The network was created using Cytoscape software. Blue and red indicate respectively the down- and up regulated proteins.
(E) Enrichment map of significant gene ontology (GO) terms between S. epidermidis and control mice. Nodes represent gene sets. Highly similar protein sets are connected by edges, grouped in sub-clusters, and annotated automatically.
(F) Top 20 significantly enriched IPA Diseases and functions (p < 0.01) sorted by Z score (red indicates positive Z score).
(G) Upstream regulator analysis showing the effects of S. epidermidis infection. Red indicates increased and green indicates decreased measurements of proteins. Orange dashed lines predict activation, blue dashed lines predict inhibition and gray dashed lines indicate a non-predicted response (see also Table S2).