Figure 2.
Genome editing of human E-selectin to attach the 11 amino acid HiBiT sequence to the N terminus
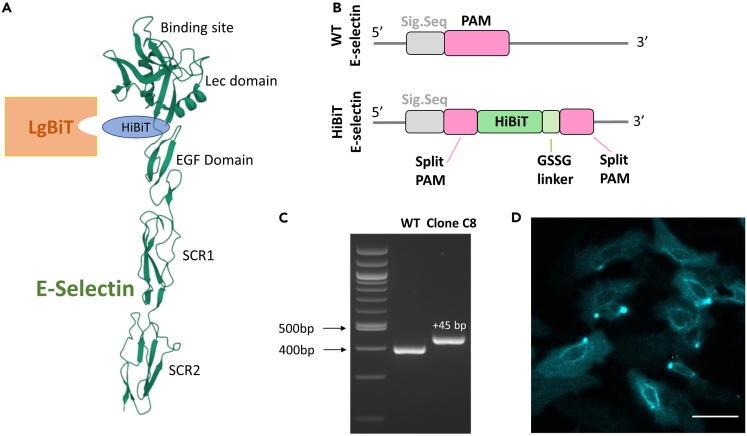
(A) Schematic of the position of HiBiT on the N terminus of E-selectin and its re-complementation with LgBiT. The E-selectin structure shown is the extended structure of the N-terminal regions of E-selectin (PDB: 4C16) showing the binding site for glycomimetic 1/sLex and the N-terminal Lectin (Lec), EGF-like and first two SCR domains.
(B) General genome-editing strategy showing the PAM sequence adjacent to the signal sequence of E-selectin and the insertion of HiBiT and a GSSG linker immediately following the signal sequence in the HiBiT-tagged E-selectin sequence. The editing disrupts the PAM sequence thus preventing any further modification by Cas9.
(C) Agarose gel (3%) of a PCR amplified 370 bp fragment of the N-terminal region of E-selectin in wild-type HUVECs and HiBiT-edited TERT2-HUVEC clone C8. Successful homozygous edit of HiBiT TERT2-HUVEC clone C8 was demonstrated by the appearance of a single band representing the 45 bp addition (HiBiT + GSSG linker) in length of the PCR fragment compared to unedited HUVECs. A 100 bp ladder is also shown for comparison.
(D) Bioluminescence imaging of HiBiT TERT2-HUVEC clone C8 cells expressing HiBiT-E-selectin under endogenous promotion. Cells were treated with 1 nM TNFα for 6 h and then incubated with purified LgBiT protein (50 nM in 0.1% BSA HBSS) for 20 min (37°C). Furimazine was then added (1:400 in 0.1% BSA HBSS) (5 min, RT) and 512 x 512 images were captured using bioluminescence filters (exposure 20 s, gain 200) on the Olympus LuminoView 200. Scale bar represents 50 μm. The image shown is representative of 5 similar experiments.