Figure 4.
Microfluidic manipulations can affect cell death and subsequent ambient contamination in downstream data
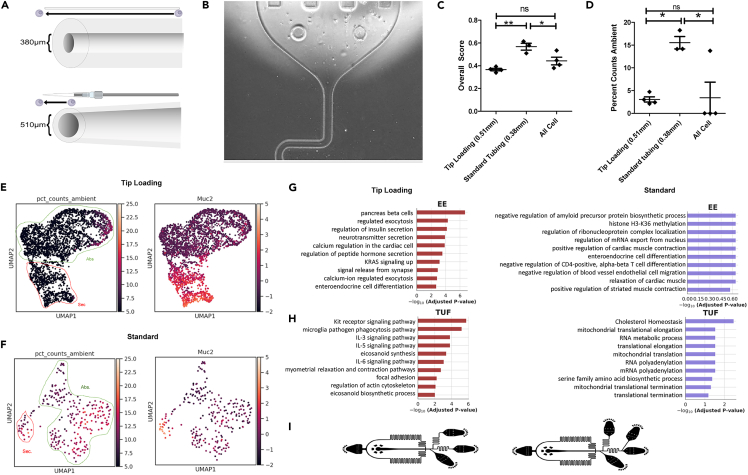
(A) Schematic of standard loading (top) and tip loading (bottom).
(B) Live hopper visualization of viable single cells from tip loading apparatus.
(C and D) Quantification of (C) AmbiQuant overall score, (D) percent counts ambient comparing various microfluidics manipulations. Mean with SEM as error bars for n = 3 or 4 samples. ∗p < 0.05, ∗∗p < 0.01 by ANOVA followed Tukey post-test.
(E and F) UMAP overlay with percent counts ambient or Muc2 expression for (E) tip loading or (F) standard loading. Secretory (red) and absorptive (green) lineages are outlined. Gene expression values on scale bars are Z-scores of normalized values described in STAR Methods.
(G and H) Comparison of functional enrichment analysis datasets derived from tip loading (higher data quality) and standard loading (lower data quality) looking at (G) enteroendocrine (EE) and (H) Tuft (TUF) cells.
(I) Schematic for standard inDrops chip (left), and All Cell chip (right).