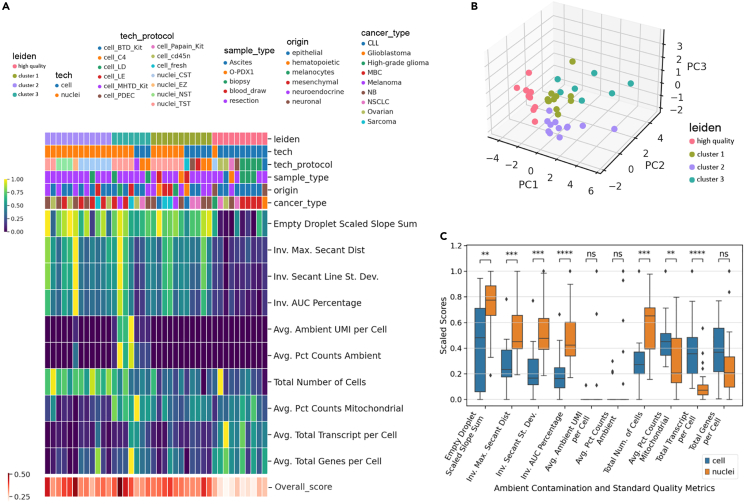
Figure 5.
Ambient contamination and quality control metrics reveal impact of intrinsic and extrinsic factors on data quality
(A) Heatmap of ambient contamination and standard QC metric scores with HTAPP datasets as columns grouped by Leiden clusters. Metrics are shown as rows, where the first ten rows are individual metrics, whose colors correspond to the top left color bar. The last row is the AmbiQuant overall score for the ambient contamination metrics, colored in red corresponding to the bottom left color bar. The Leiden cluster labels and labels of isolation technique, technique and protocol combination, sample type, tissue origin, and cancer type are shown as color bars above the heatmap. Metric scores are normalized between 0 and 1 for each row for visualization. Abbreviations - cell: scRNA-seq; nuclei: snRNA-seq; BTD: brain tumor dissociation; C4: collagenase 4 and DNase I; LD: Liberase TM and DNase I; LE: Liberase TM, elastase and DNase I; Miltenyi Biotec human tumor dissociation; PDEC: pronase, dispase, elastase, collagenases A and 4 and DNase I; Paipan: (cysteine protease); cd45n: CD45+ depletion; CST: CHAPS with salts and Tris; EZ: EZPrep; NST: Nonidet P40 with salts and Tris; TST: Tween with salts and Tris; O-PDX1: orthotopic patient-derived xenograft; CLL: Chronic lymphocytic leukemia; MBC: Metastatic breast cancer; NB: Neuroblastoma; NSCLC: Non-small cell lung carcinoma.
(B) Three-dimensional scatterplot of the first 3 principal components of the ambient contamination and standard QC metric score matrix colored by Leiden cluster labels.
(C) Boxplot comparing the metric scores between single-cell and single-nucleus sequenced samples. Two-sided Mann-Whitney-Wilcoxon test performed between single-cell and single-nuclei groups. ∗∗p < 0.01, ∗∗∗p < 0.001,∗∗∗∗p < 0.0001.