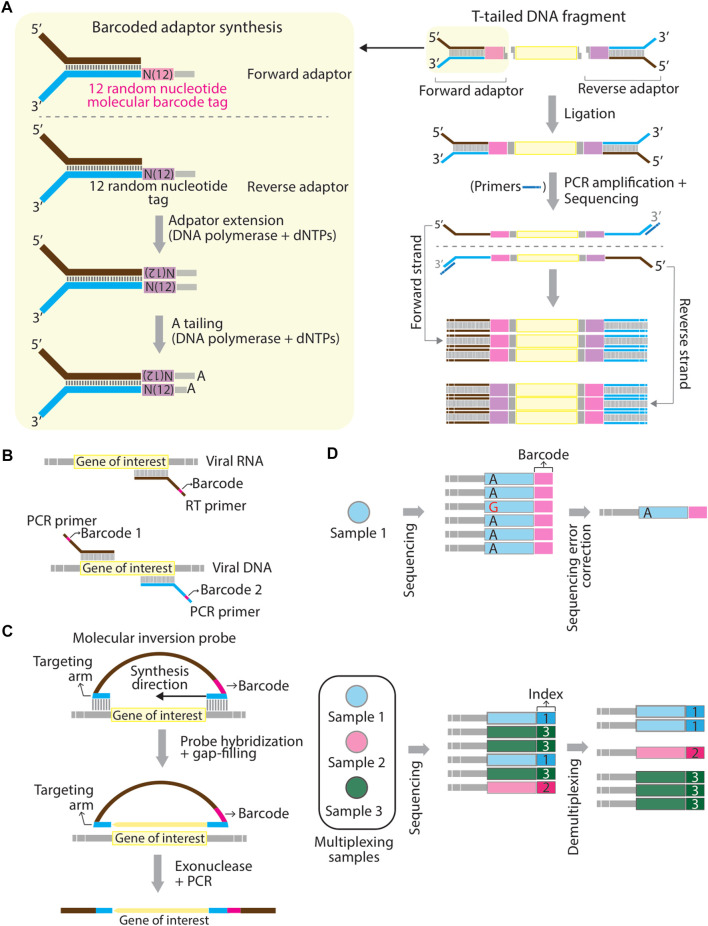
FIGURE 1.
Schematic representation of mechanistic strategies of barcoding. (A–C) Barcodes can be introduced to a template using adaptors through direct ligation (A), using RT- or PCR primers at the reverse transcription or PCR amplification step (B), and using hybridizing molecular inversion probes (C). (D) Schematic representation of the difference between “barcodes” and “sample indexes”. Barcodes aim to correct sequencing errors. For example, a misreading nucleotide, guanosine (G) can be corrected in final consensus sequences for a pool of Sample 1 (top panel). Sample indexes are used to multiplex different sequencing amplicons generated from different pools of samples (Sample 1, 2, and 3) (bottom panel). Panel (A) is modified based on Figure 1 in (Schmitt et al., 2012) and panel (C) is modified based on Figure 1 in (Hiatt et al., 2013).