Table 2.
Molecular docking/binding energy score for all derivatives represented.
| Sl. No. | PubChem CID | Chemical structure | Lassa virus glycoprotein spike (PDB ID 5FT2) | Lassa virus nucleoprotein (PDB ID 3MX5) |
|---|---|---|---|---|
| Binding affinity (kcal/mol) | Binding affinity (kcal/mol) | |||
| 01 | Evodiamine |
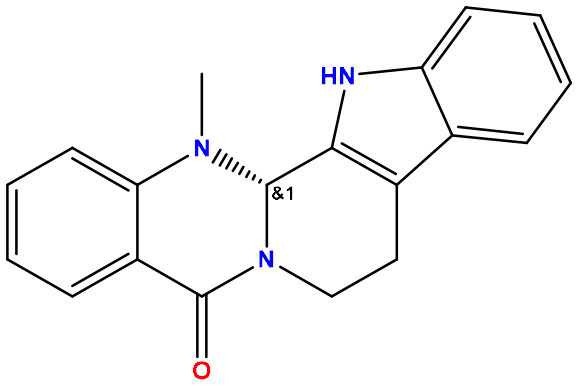
|
−7.2 | −10.7 |
| 02 | 49806754 |

|
−7.4 | −9.1 |
| 03 | 49806624 |
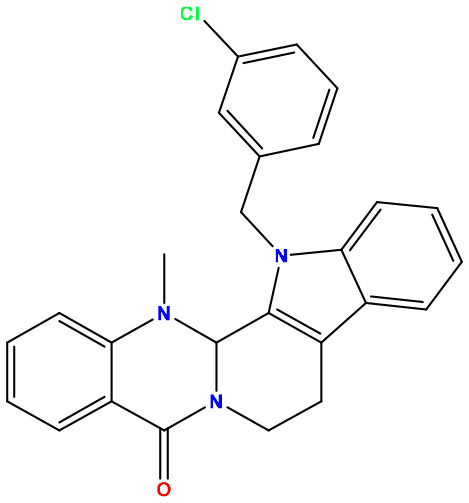
|
−8.3 | −9.7 |
| 04 | 49806625 |
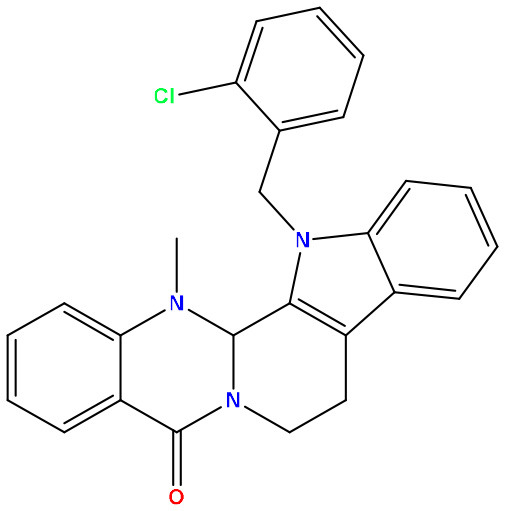
|
−8.5 | −11.0 |
| 05 | 49806500 |

|
−8.1 | −9.8 |
| 06 | 129710532 |

|
−1.7 | −2.0 |
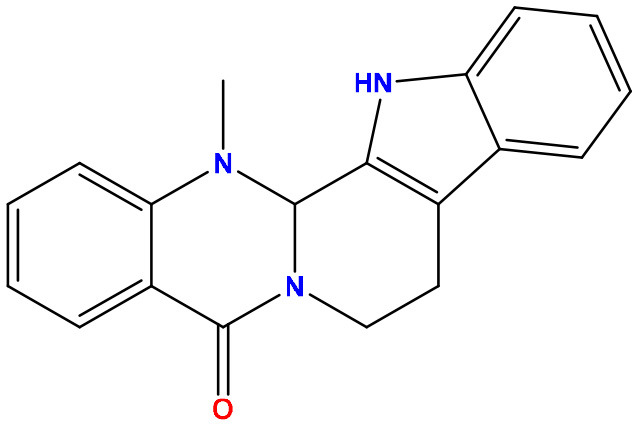
| 07 | 151289 |
|
−7.8 | −10.6 |
| 08 | 56967508 |

|
−7.2 | −9.4 |
| 09 | 49804912 |

|
−8.3 | −10.3 |
| Standard Sofosbuvir |
|
−5.9 | −7.1 |
