Abstract
Background:
Glioma is one of the most malignant and aggressive tumors, with an extremely poor prognosis. Human telomerase reverse transcriptase (hTERT) promoter mutation is regarded as a risk factor in tumor growth. Although the prevalence of hTERT promoter (pTERT) mutation in gliomas has been investigated, the results are inconsistent. This meta-analysis aims to investigate the prognostic value of hTERT in glioma patients and its interaction with other biomarkers.
Materials and Methods:
We searched 244 citations from four databases: PubMed (2000–2021), Web of Science (2000–2021), Embase (2010–2021), and Cochrane Library (2000–2021) with 28 articles included.
Results:
We calculated hazard ratios (HRs) using the random effect model and the pooled result suggested that TERT promoter mutation predicted poorer overall survival (HR: 1.53, 95% confidence interval [CI]: 1.34–1.75, P < 0.001, I2: 49.9%, pheterogeneity:0.002) and progression-free survival (HR: 1.55, 95% CI: 1.27–1.88, P < 0.001, I2: 0.0%, pheterogeneity: 0.473). For subgroup analysis, we analyzed multiple factors including iso-citrate dehydrogenase (IDH) genotype, age, diagnosis, pTERT region, so as to locate the sources of heterogeneity. Interestingly, in IDH mutant subgroup, pTERT mutation became a beneficial prognostic factor (HR: 0.73, 95% CI: 0.57–0.93, I2: 22.3%, pheterogeneity: 0.277), which is contrary to the results in pooled analysis.
Conclusion:
In general, pTERT mutation may result in shorter survival time in glioma patients, but longer survival time when glioma patients are combined with IDH mutation.
Keywords: Glioma, human telomerase reverse transcriptase, iso-citrate dehydrogenase, meta-analysis, prognosis
INTRODUCTION
There is an urgent need to address the healthy problems caused by glioma, a malignant tumor in central nervous system, which is a growing public health concern worldwide for its 40% of primary brain tumors accompanied by a poor prognosis. Gliomas can be classified into astrocytic, oligodendroglial, and other neuroepithelial tumors, of which glioblastoma (GBM) has the worst prognosis and is most common in primary brain tumors.[1,2] A growing body of literature hint at the importance of the telomerase reverse transcriptase (human telomerase reverse transcriptase [hTERT]) in oncogenesis. In broad terms, hTERT is understood as a reverse transcriptase with high specificity that adds repeats to 3’ end of chromosomes with key role in maintaining chromosomes integrity,[3] which has a sharp positive correlation between repeated cell replication and oncogenesis.[4] Tremendous research has been published, suggesting pTERT mutation in glioma patients, with mainly two types of mutation in hTERT promoter (pTERT) region: C228T and C250T,[4] desired molecules into a new binding section for transcriptional enhancers,[4] which in turn upregulates TERT. Since TERT expression in body cells is extremely low, a pTERT mutation is generally thought to be a hallmark of oncogenesis. Recently, a considerable amount of literature has been published suggesting a worse survival in pTERT mutated glioma patients. However, interestingly, some studies indicated differently with adverse ratios (hazard ratio [HR] <1) in their results. Due to this inconsistency, the aim of our research is to conduct a meta-analysis seeking for the correlation of pTERT mutation and survival of glioma patients, which contribute to prognosis and develop targeted drug that can lead to new curative therapies.
METHODS
Searching strategy
Pertinent publications that concentrate on the subject of glioma more frequently adopt a historical or chronological approach were carried out based on four databases: PubMed, Web of Science, Embase, and Cochrane Library from 2000 to January 26, 2022. The studies were restricted to humans and there were no place and publication status limitations. The broad use of the key term “glioma,” “hTERT,” and “prognosis” is sometimes broadened to include (hTERT) AND (glioma OR glial cell tumor OR astrocytoma OR GBM) AND (prognosis OR prognostic OR prognoses). The search volume with high correlation on databases and some articles related to the searched keywords was also viewed for potential studies. The functional information of regularly imported data was all from the electronic databases into EndNote with duplicates deleted. Articles were independently assessed by two reviewers (Rongxuan Hua and Qiuxuan Li) using predesigned eligibility forms, according to defined inclusion criteria and exclusion criteria. Any disagreement between investigators was resolved by consensus.
Inclusion and exclusion criteria
Although extensive research has been carried out on glioma, no single study has analyzed the complex clinical manifestations and possible mechanisms. Analysis was based on the conceptual framework proposed by following standards listed as follows: First, there are investigations on TERT promoter mutation in glioma patients; second, they evaluated the survival outcomes including overall survival (OS) or progression-free survival (PFS) in glioma patients; third, HRs and 95% confidence interval (CI) are available or able to be calculated with enough data. Last but not least, the studies about TERT expression affects the survival rate in glioma patients were also taken into consideration.
Exclusion were as follows: (i) are about other tumors but glioma; (ii) without successive records of following-up; (iii) were no enough data for HRs and 95% CIs calculation; (iv) the data are from neither clinical nor experimental studies, but from other sources such as reviews, posters, conference papers, letters, etc.; and (v) a clear benefit of factor in the prevention of glioma could not be identified in this analysis.
Quality assessment
To evaluate the quality and methodological appropriateness of included articles, provide guidance for future research as well, a wide variety of relevant parameters for disease evaluation have been proposed, including the Newcastle–Ottawa Scale in the meta-analysis.[5] Two reviewers independently awarded stars and studies can receive a maximum score of nine stars. Studies awarded more than five stars regarded to be of high-quality and low-quality are awarded a star.
Data extraction
All data were extracted using Microsoft Office Excel 2019 as dichotomous outcomes (primary outcome: OS and secondary outcome: PFS). All of the content related to the research is extracted, including the names of the first author, publication years, ethnicity, diagnosis group, stages of tumors, sample size, numbers of male and female, mean age, cutoff values, outcomes, HRs combined with 95% CIs, therapy methods, variable types, and investigation design including prospective and retrospective studies to corroborate the initial diagnosis and study designs (retrospective or prospective). For several studies that did not offer HRs and 95% CIs, we extracted them from Kaplan − Meier curves in original articles. In order to make the obtained data have more objectivity and accuracy, all the related data were extracted by two independent authors (Rongxuan Hua and Qiuxuan Li) and discussed with a third author (Han Gao) if disagreement occurs.
Statistical analysis
HRs and 95% CIs were directly extracted from included articles if the data was straightly given, if not, Engauge Digitizer Software version 4.1 (Mark Mitchell, Baurzhan Muftakhidinov and Tobias Winchen et al.URL: http://markummitchell.github.io/engauge-digitizer) was used for retrieving the HRs and the 95% CIs from the Kaplan-Meier curves. When a HR >1, it indicates a worse prognosis in glioma patients with mutant pTERT. StataSE version 12.0 (Stata Corp, College Station, TX, USA) was used for the following statistical analyses. HRs and 95% CIs were combined in analyzing the association of pTERT mutation with the prognosis of glioma patients. Cochran's Q test and Higgins I2 statistic were conducted to evaluate the heterogeneity. For Pheterogeneity <0.10 or I2 >50% in the literature,[6] which indicates a significant heterogeneity, we use random-effect model (DerSimonian–Laird method). Otherwise, the fixed-effects model (Mantel–Haenszel method) was undertaken. Subgroup analysis was conducted to investigate the sources of heterogeneity among included studies. We separately categorized 28 studies into Subgroup 1-6 based on following stratification: iso-citrate dehydrogenase (IDH) genotype (wildtype/mutant), age (adult/adult and children), diagnosis (astrocytoma/GBM), pTERT region (C228T/C250T), ethnicity (Asian/Caucasian), and TERT evaluation type (promoter mutation/protein expression). Publication bias was assessed by Begg's funnel. Sensitivity analysis was conducted for locating sensitive factor among the included studies. During statistical analysis, exponential transformation has been done. The statistically significant threshold is set to P < 0.05.
RESULTS
Study characteristics
The initial search strategies retrieved 244 articles with 72 duplicates. One hundred and seventy-two articles were identified for title and abstract screening, 113 of them were excluded for three reasons. Afterward, 59 of them were taken into full-text evaluation with 31 further excluded. Eventually, 28 articles were included for our meta-analysis.[7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34] Among them, 21 studies[7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27] were about pTERT mutation and others[28,29,30,31,32,33,34] were about TERT expression [Figure 1].
Figure 1.
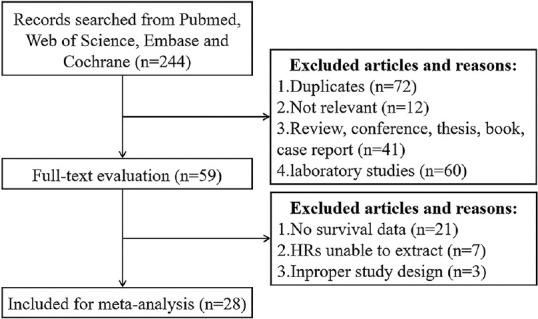
Flow diagram presenting the data selection process and criterions followed in this meta-analysis
This system of selection has been broadened to include 28 articles (2013–2021) according to whether meet the inclusion standards discussed detail above. Each study has a sample size range from 20 to 807 with a total of 5881 participants, which came from 13 countries. By way of illustration, approximately 75% those surveyed focus more attention on pTERT mutational, which is optimized as a whole meta-analysis. The remaining 25% of the literature is concerned with the expression of TERT, which, as a parameter, can only be managed a subgroup analysis. Particular variables have been extracted including ethnicity, cancer type, sample size, age of patients, cutoff values, etc., Those factors were not all integrated, although we have read every study thoroughly; therefore, we put only those who have certain data into consideration during each subgroup analysis.
To assess whether and how glioma is produced and received, the genotyping was measured by reverse transcription–polymerase chain reaction (RT-PCR), real time-PCR, and PCR-Sanger sequencing. In general, strong evidence suggests a positive correlation between the pTERT mutation (or TERT expression) and OS in glioma. All mentioned characteristics of the study are listed in Tables 1-3.
Table 1.
Characteristics of 1-10 studies included in the meta-analysis
| Author (year) | Ethnicity | Cancer type | Sample size | Age | Cutoff (hTERT) | Outcome | R | 95% CI | NOS score | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Lower | Upper | |||||||||
| Gramatzki (2021) | Caucasian | GBM (IV) | 298 | Adult | Mutant versus wildtype | OS | 1.74 | 1.07 | 2.82 | 7 |
| Razis (2020) | Caucasian | Gliomas (III-IV) | 77 | Adult | Mutant versus wildtype | OS | 1.88 | 1.00 | 3.51 | 8 |
| Mutant/IDH wildtype versus wildtype/IDH wildtype | OS | 2.26 | 1.09 | 4.66 | ||||||
| Cheng (2020) | Asian | Gliomas (II-IV) | 395 | All | Mutant versus wildtype | OS | 1.44 | 0.93 | 2.23 | 7 |
| Kessler (2020)* | Caucasian | Primary GBM (IV) | 455 | NA | Mutant (C228T) versus wildtype | OS | 1.23 | 0.77 | 1.97 | 7 |
| Mutant (C250T) versus wildtype | OS | 2.56 | 1.2 | 5.44 | ||||||
| Recurrent GBM (IV) | Mutant (C228T) versus wildtype | OS | 0.80 | 0.54 | 1.20 | |||||
| Mutant (C250T) versus wildtype | OS | 1.34 | 0.89 | 2.01 | ||||||
| Tamrakar (2019) | Asian | Gliomas (II-IV) | 97 | All | Mutant versus wildtype | OS | 1.20 | 0.50 | 2.80 | 7 |
| Kuwahara (2019) | Asian | Astrocytoma (II-III) | 36 | Adult | Mutant versus wildtype | OS | 12.5 | 2.17 | 100.00 | 7 |
| Kim (2018) | Asian | Gliomas (II-IV) | 67 | Adult | Mutant versus wildtype | OS | 1.055 | 0.495 | 2.422 | 8 |
| PFS | 1.436 | 0.670 | 3.080 | |||||||
| Nguyen (2017) | Caucasian | Gliomas (II-IV) | 303 | NA | Mutant/IDH wildtype versus wildtype/IDH wildtype | OS | 1.68 | 1.13 | 2.50 | 6 |
| PFS | 1.57 | 1.05 | 2.36 | |||||||
| Lee (2017) | Asian | Anaplastic Astrocytoma (III) | 67 | Adult | Mutant/IDH wildtype versus wildtype/IDH wildtype | OS | 1.110 | 0.116 | 10.608 | 8 |
| GBM (IV) | OS | 2.111 | 1.016 | 4.389 | ||||||
| Li (2017)* | Asian | Gliomas (II-IV) | 47 | All | Mutant versus wildtype | OS | 2.06 | 0.40 | 10.56 | 6 |
| Mutant versus wildtype | PFS | 2.21 | 0.92 | 5.32 | ||||||
*Data extracted form survival curve. Data of hTERT promoter mutation (n=22) and hTERT expression difference (n=7) in glioma patients. GBM=Glioblastoma; hTERT=Human telomerase reverse transcriptase; IDH=Iso-citrate dehydrogenase; PFS=Progression-free survival; OS=Overall survival; NA=Not available; CI=Confidence interval; NOS=Newcastle-Ottawa Scale
Table 3.
Characteristics of 19-28 studies included in the meta-analysis
| Author (year) | Ethnicity | Cancer type | Sample size | Age | Cutoff (hTERT) | Outcome | HR | 95% CI | NOS score | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Lower | Upper | |||||||||
| Chen C (2014) | Asian | Gliomas (III-IV) | 62 | Mutant versus wildtype | OS | 4.148 | 1.973 | 8.721 | 7 | |
| Astrocytoma (II) | OS | 3.058 | 1.886 | 4.958 | ||||||
| Killela PJ (2014)* | Caucasian | Gliomas (II) | 442 | NA | Mutant/IDH mutant versus wildtype/IDH mutant | OS | 0.93 | 0.37 | 2.37 | 9 |
| Gliomas (III) | Mutant/IDH mutant versus wildtype/IDH mutant | OS | 0.87 | 0.42 | 1.79 | |||||
| Mutant/IDH wildtype versus wildtype/IDH wildtype | OS | 1.39 | 0.27 | 7.16 | ||||||
| Gliomas (IV) | Mutant/IDH wildtype versus wildtype/IDH wildtype | OS | 1.26 | 0.87 | 1.82 | |||||
| Mutant/IDH mutant versus wildtype/IDH mutant | OS | 0.85 | 0.58 | 1.25 | ||||||
| Nonoguchi N (2013) | Caucasian | GBM (IV) | 187 | NA | Mutant (C228T) versus wildtype | OS | 1.50 | 1.07 | 2.11 | 8 |
| Mutant (C250T) versus wildtype | OS | 1.17 | 0.76 | 1.80 | ||||||
| Gandhi P (2021) | Caucasian | Gliomas (II-IV) | 135 | NA | Plasma protein expression (1.309 ng/L) | OS | 1.230 | 1.075 | 1.408 | 7 |
| Elsers D (2021) | Caucasian | GBM (IV) | 53 | Adult | IHC expression score | OS | 0.691 | 0.310 | 1.549 | 7 |
| Potharaju M (2019) | Caucasian | GBM (IV) | 87 | Adult | IRS1-6 versus IRS7-12 | OS | 1.65 | 0.94 | 2.89 | 6 |
| Rute JM (2017) | Caucasian | GBM (IV) | 55 | Adult | Expression | OS | 1.034 | 1.001 | 1.067 | 6 |
| Gandhi P (2017) | Caucasian | Gliomas (II-IV) | 72 | NA | Intra-tumor expression 2.315% | OS | 1.057 | 1.030 | 1.080 | 6 |
| Plasma protein 1.309 ng/L | OS | 1.322 | 1.160 | 1.510 | ||||||
| Dorris K (2014)* | Caucasian | Gliomas (IV) | 20 | Adult | Protein expression | OS | 0.90 | 0.04 | 19.25 | 6 |
| Lötsch D (2013) | Caucasian | GBM (IV) | 100 | All | mRNA expression | OS | 1.02 | 0.61 | 1.71 | 7 |
*Data extracted form survival curve. Data of hTERT promoter mutation (n=22) and hTERT expression difference (n=7) in glioma patients. GBM=Glioblastoma; hTERT=Human telomerase reverse transcriptase; IDH=Iso-citrate dehydrogenase; OS=Overall survival; NA=Not available; CI=Confidence interval; NOS=Newcastle-Ottawa Scale; HR=Hazard ratio; IHC=Immunohistochemistry; IRS=Immunoreactivity score
Table 2.
Characteristics of 11-18 studies included in the meta-analysis
| Author (year) | Ethnicity | Cancer type | Sample size | Age | Cutoff (hTERT) | Outcome | HR | 95% CI | NOS score | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Lower | Upper | |||||||||
| Nencha U (2016)* | Caucasian | GBM (IV) | 651 | NA | Mutant versus wildtype | OS | 1.46 | 1.19 | 1.80 | 7 |
| Gao K (2015) | Asian | Gliomas (I-IV) | 389 | Adult | Mutant versus wildtype | OS | 1.39 | 1.13 | 2.17 | 7 |
| Mutant (C228T) versus wildtype | OS | 7.66 | 2.21 | 26.61 | ||||||
| Mutant (C250T) versus wildtype | OS | 2.5 | 0.57 | 10.91 | ||||||
| Zhang Z (2015) | Asian | Gliomas (II-III) | 295 | NA | Mutant versus wildtype | OS | 1.158 | 0.746 | 1.796 | 7 |
| PFS | 1.139 | 0.756 | 1.716 | |||||||
| Mutant/IDH mutant versus wildtype/IDH mutant | OS | 0.890 | 0.349 | 2.267 | ||||||
| Spiegl-Kreinecker S (2015) | Caucasian | GBM (IV) | 126 | NA | Mutant versus wildtype | OS | 1.683 | 1.048 | 2.703 | 8 |
| Simon M (2015) | Caucasian | GBM (IV) | 176 | Adult | Mutant versus wildtype | OS | 2.05 | 1.30 | 3.23 | 7 |
| Mosrati MA (2015) | Caucasian | GBM (IV) | 146 | NA | Mutant (C228T) versus wildtype | OS | 4.04 | 1.55 | 10.51 | 7 |
| Mutant (C250T) versus wildtype | OS | 3.7 | 1.3 | 10.51 | ||||||
| Labussière M (2015) | Caucasian | Glioms (II-IV) | 807 | Adult | Mutant versus wildtype | OS | 1.497 | 1.071 | 2.092 | 8 |
| PFS | 1.766 | 1.299 | 2.401 | 9 | ||||||
| Mutant/IDH mutant versus wildtype/IDH mutant | OS | 0.57 | 0.32 | 1.00 | ||||||
| Chan AK (2015)* | Asian | Gliomas (II-III) | 236 | NA | Mutant/IDH mutant versus wildtype/IDH mutant | OS | 0.46 | 0.24 | 0.88 | 9 |
| Mutant/IDH wildtype versus wildtype/IDH wildtype | OS | 2.70 | 1.36 | 5.37 | ||||||
| Astrocytoma (II-III) | Mutant/IDH wildtype versus wildtype/IDH wildtype | OS | 2.42 | 1.14 | 5.15 | |||||
*Data extracted form survival curve. Data of hTERT promoter mutation (n=22) and hTERT expression difference (n=7) in glioma patients. GBM=Glioblastoma; hTERT=Human telomerase reverse transcriptase; IDH=Iso-citrate dehydrogenase; PFS=Progression-free survival; OS=Overall survival; NA=Not available; CI=Confidence interval; NOS=Newcastle-Ottawa Scale; HR=Hazard ratio
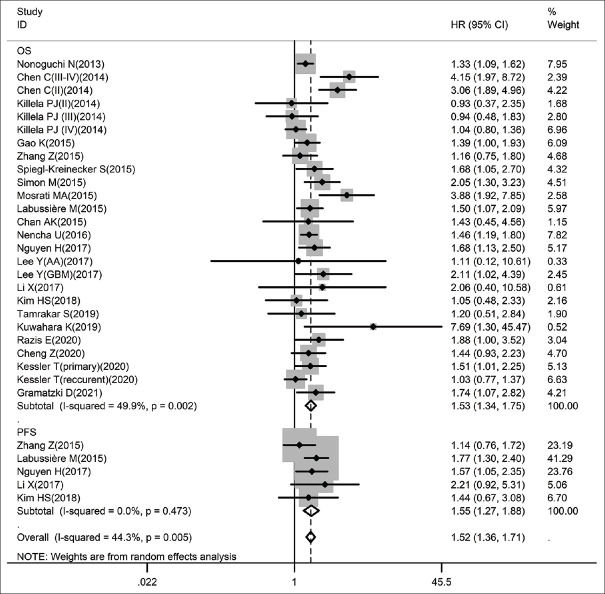
Influences of pTERT mutation on overall survival and progression-free survival in glioma patients
Figure 2 presents the results of pooled analysis among 21 studies measures of the data of pTERT mutation and OS in glioma participants. In order to confirm the positive correlation between pTERT mutation and OS/PFS of glioma by meta-analysis, the random-effect model is one of the more practical ways to capture the complexities of the phenomenon of bases on the data with obviously heterogeneity (I2 = 49.9%, P = 0.002). What can be clearly seen in this Figure 2 is the dramatic positive correlation was found between the mutated pTERT and a worse outcome for both OS (combined HR: 1.53, 95% CI: 1.34–1.75, P < 0.001, I2: 49.9%, pheterogeneity: 0.002) and PFS (combined HR: 1.55, 95% CI: 1.27–1.88, P < 0.001, I2: 0.0%, pheterogeneity: 0.473) in glioma patients.
Figure 2.
Forest plots for studies on the association between pTERT mutation and OS/PFS in glioma among 28 studies included in the meta-analysis (Some contain multiple HRs). Results are presented as individual and pooled HR (95% CI). OS: Overall survival, PFS: Progression-free survival, HR: Hazard ratio, CI: Confidence interval
Subgroup analysis
Given the significant heterogeneity in this meta-analysis of 25% of all the included articles, the reliability of measures of subgroup analyses was assessed to better understand the heterogeneity. The participants were stratified into subgroup 1–6 separately by: IDH genotype, age, diagnosis, pTERT region, ethnicity, and TERT evaluation type.
Further analysis subgroup 1 based on IDH genotype provides the significant positive correlation between pTERT mutations and worse OS in IDH-wildtype gliomas [HR: 1.69, 95% CI: 1.36–2.10, I2 = 26.1%, pheterogeneity: 0.247; Figure 3]. Next, we found a most striking observation that the pTERT mutation became a benefit factor in IDH-mutant patients [HR: 0.73, 95% CI: 0.57–0.93, I2 = 22.3%, pheterogeneity: 0.277; Figure 4], which greatly explain the heterogeneity in pooled analysis. To gain further insights, we next collected additional data in more detail from other subgroups to illustrate two potential indexes. Subgroup 2: The results, as shown in Table 4, indicate that a clear disadvantage of pTERT mutation in OS of astrocytoma patients (HR: 2.91, 95% CI: 1.97–4.31, I2 = 0.00%, pheterogeneity: 0.548) than in GBM patients (HR: 1.54, 95% CI: 1.28–1.85, I2 = 49.8%, pheterogeneity: 0.021). Subgroup 3: This result is somewhat counterintuitive that pTERT mutation is slightly more unfavorable in studies that included only adults (HR: 1.62, 95% CI: 1.36–1.92, I2 = 0.00%, pheterogeneity: 0.451) rather than that included both adults and children (HR: 1.42, 95% CI: 0.97–2.07, I2 = 0.00%, pheterogeneity: 0.840) in prognosis. Subgroup 4: Stratification by pTERT mutation region found that both C228T (HR: 1.72, 95% CI: 1.00–2.97, I2 = 79.6%, pheterogeneity: 0.001) and C250T (HR: 1.50, 95% CI: 1.15–1.95, I2 = 41.4%, pheterogeneity: 0.146) mutation are negative predictor for OS in glioma. Subgroup 5: Stratification by ethnicity shows that pTERT as an unfavorable factor in Asian (HR: 1.65, 95% CI: 1.35–1.80, I2 = 51.7%, pheterogeneity: 0.010) and Caucasian (HR: 1.49, 95% CI: 1.28–1.74, I2 = 56.4%, pheterogeneity: 0.014) glioma patient. Subgroup 6: The data demonstrated that for TERT evaluation type, pooled HR was 1.56 (95% CI: 1.35–1.80, I2 = 57.2%, pheterogeneity: 0.001) for pTERT mutation; and 1.11 (95% CI: 1.04–1.18, I2 = 67.1%, pheterogeneity: 0.003) for TERT expression.
Figure 3.
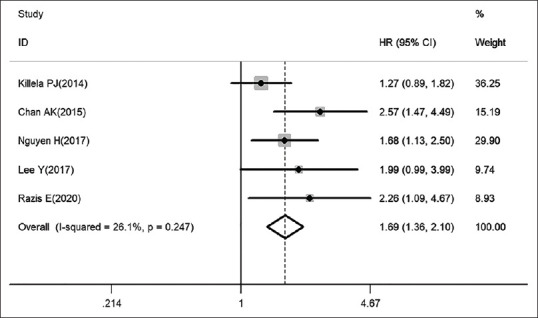
PTERT mutation associated with worse prognosis in IDH-wildtype patients (fixed-effect model). IDH: Iso-citrate dehydrogenase
Figure 4.
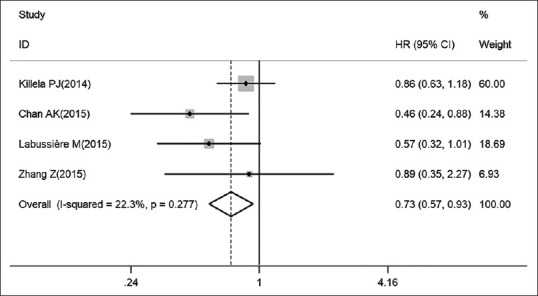
PTERT mutation leads to pleasant prognosis outcomes in IDH-mutant glioma patients (fixed-effect models). IDH: Iso-citrate dehydrogenase
Table 4.
Summary of the subgroup meta-analysis results
| Analysis (OS) | n | References | Fixed-effect model | Random-effect model | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| HR (95% CI) | P | HR (95% CI) | P (Z-test) | I2 (%) | Ph | |||
| Subgroup 1 | ||||||||
| IDH-wildtype | 5 | [8,14,16,23,25] | 1.69 (1.36-2.10) | 0.000 | 1.76 (1.42-2. 17) | 0.000 | 22.5 | 0.271 |
| IDH-mutant | 4 | [19,23,24,25] | 0.73 (0.57-0.93) | 0.012 | 0.70 (0.52-0.95) | 0.023 | 22.3 | 0.277 |
| Subgroup 2 | ||||||||
| GBM | 8 | [7,10,16,17,20,21,22,27] | 1.47 (1.31-1.65) | 0.000 | 1.54 (1.28-1.85) | 0.000 | 49.8 | 0.021 |
| Astrocytoma | 4 | [12,16,23,26] | 2.97 (1.97-4.31) | 0.000 | 2.91 (1.97-4.31) | 0.000 | 0.00 | 0.548 |
| Subgroup 3 | ||||||||
| Adult | 8 | [7,8,12,13,16,18,21,24] | 1.62 (1.36-1.92) | 0.000 | 1.62 (1.36-1.92) | 0.000 | 0.00 | 0.451 |
| Adult and children | 3 | [9,11,15] | 1.42 (0.97-2.07) | 0.072 | 1.42 (0.97-2.07) | 0.072 | 0.00 | 0.840 |
| Subgroup 4 | ||||||||
| C228T | 4 | [10,18,22,27] | 1.32 (1.06-1.64) | 0.012 | 1.72 (1.00-2.97) | 0.052 | 79.6 | 0.001 |
| C250T | 4 | [10,18,22,27] | 1.50 (1.15-1.95) | 0.003 | 1.67 (1.13-2.47) | 0.010 | 41.4 | 0.146 |
| Subgroup 5 | ||||||||
| Asian | 10 | [9,11,12,13,15,16,18,19,23,26] | 1.64 (1.38-1.96) | 0.000 | 1.65 (1.35-1.80) | 0.001 | 51.7 | 0.010 |
| Caucasian | 11 | [7,8,10,14,17,20,21,22,24,25,27] | 1.39 (1.27-1.52) | 0.000 | 1.49 (1.28-1.74) | 0.000 | 56.4 | 0.014 |
| Subgroup 6 | ||||||||
| pTERT mutation | 21 | [7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27] | 1.44 (1.33-1.56) | 0.000 | 1.56 (1.35-1.80) | 0.000 | 57.2 | 0.001 |
| TERT expression | 7 | [28,29,30,31,32,33,34] | 1.06 (1.04-1.08) | 0.000 | 1.11 (1.04-1.18) | 0.001 | 67.1 | 0.003 |
n=Number of studies; Ph=P values of Q-test for heterogeneity test. HR=Hazard ratio; CI=Confidence interval; OS=Overall survival; TERT=Telomerase reverse transcriptase; GBM=Glioblastoma; IDH=Iso-citrate dehydrogenase
In general, through subgroup analysis, we may draw a conclusion that IDH mutation is an unneglectable factor in the association of TERT and prognosis. Moreover, TERT mutation seems to be a worse risk factor in astrocytoma patients. In addition, it is inappropriate to take both children and adults into accounts, since our results showed no significant differences (P > 0.05) in those studies that did not exclude children.
Publication bias and sensitivity analysis
No evidence for notable publication bias for the analysis between pTERT mutation and OS [Pr> |z| = 0.065; Figure 5] or PFS [Pr> |z| = 0.806; Figure 6] was found in Begg's funnel test. Meanwhile, sensitivity analysis suggested that none of the studies affected the combined HR for OS significantly [Figure 7]. Because of the amount of studies on TERT expression was not adequate enough, we did not test its publication bias and sensitivity analysis.
Figure 5.
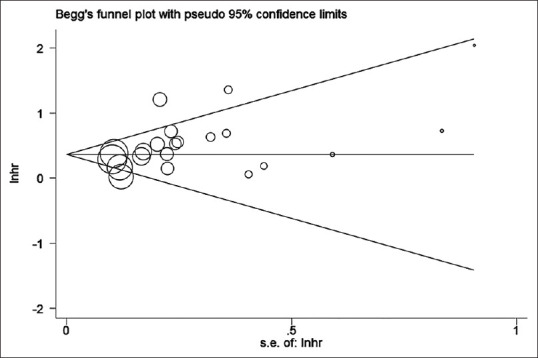
Begg's funnel plot testing publication bias in studies reporting association of OS and pTERT mutation; P = 0.065. OS: Overall survival
Figure 6.
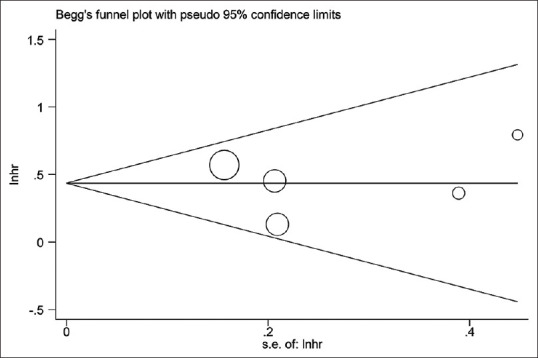
Begg's funnel plot testing publication bias in studies reporting association of PFS and pTERT mutation; P = 0.806. PFS: Progression-free survival
Figure 7.
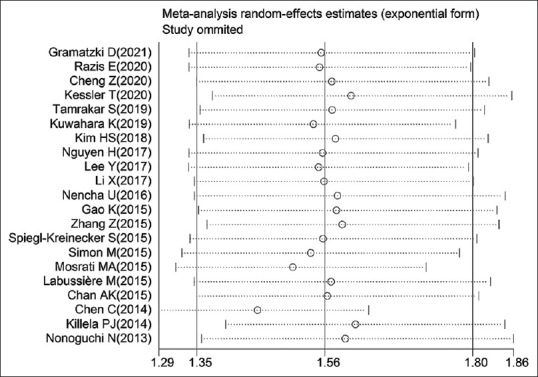
Sensitivity analysis investigating whether certain study remarkably affected the combined HR for OS (with random-effect model). OS: Overall survival, HR: Hazard ratio
DISCUSSION
As a key factor in tumor oncogenesis, multiple regression analysis revealed that TERT plays an important part in maintaining chromosomes integrity, allowing tumor cell to replicate repeatedly thus regulating the growth of cancer.[35] pTERT promoter mutation is beneficial for its expression, thus may function as a risk factor in tumor. Recently, molecular factors including pTERT, IDH, MGMT (O6-methylguanine-DNA methyltransferase) promoter methylation,[36] Chromosome 1p/19q co-deletion,[37] etc., have found their rising status in disease classification and diagnosis due to the limitation of histological classification. That means TERT is one of the hotspots which we are trying to obtain a full understanding in cancers, especially gliomas. Recent studies on glioma have investigated in TERT, as well as its correlated molecules, which contributes to the rapidly expanding field of comprehensive investigation, and found that TERT promoter mutation may have a negative impact on glioma patients’ prognosis. Surprisingly, some studies suggested patients with IDH mutation achieve the best survival rate when TERT promoter co-mutated, even better than IDH mutation alone, which was thought to be a protective factor in glioma patients’ survival.[38] However, in glioma patients, its inconsistent with previous results of survival make it harder to be applied in diagnosing and targeted drug using. These differences can be explained in part by the proximity of the SNP on TERT and the differences among individuals.[39] Some investigations have also suggested that TERT may have an impact on adjuvant therapies and radiotherapy resistance,[18,19] which makes it more significant and urgent for confirming the role that TERT plays in glioma. That's why there is abundant room for further progress in conducting the meta-analysis as an important issue for present research.
The cohort in our meta-analysis included 28 studies from 2013 to 2021, with 5881 patients in gliomas from 13 different countries involved according to the inclusion criteria. We used the HRs and 95% CIs reported in those studies as many as we could for pooled analysis and subgroup analysis, and those who didn’t provide adequate HRs and 95% CIs, we extracted them from Kaplan–Meier curves. Included studies were divided and analyzed separately into studies focused on TERT promoter mutation and TERT expression. Because of the insufficient results in the latter, we did not conduct them in pooled analysis, publication bias test, and sensitivity analysis.
Results suggesting hTERT promoter mutation significantly predicted poorer survival in patients with glioma. Patients with mutated pTERT are more likely to have shorter OS and PFS after surgery or treatment. We also found the correlation between elevated TERT expression and worse OS in glioma. However, there are heterogeneity in our meta-analysis which we believed is caused by many factors such as: most of the study data have not separate pTERT mutation region (C228T and C250T), few of them take IDH mutation into consideration, the patients had quite different age and stages. Luckily, some of the studies have clearer data on those factors mentioned above, so we next conducted subgroup analysis for further examination. Results showed a significant difference between IDH mutation and wild type patients (subgroup 1): patients who have mutated IDH genotype are expected to have longer survival through pTERT mutation, which suggested the opposite since pooled HR showed pTERT mutation as a risk factor. Meanwhile, there are several limitations in this study that need to be mentioned. First, this study is fully retrospective, thus there will inevitably be a subjective bias. Second, the articles we searched were all in the English language, which may result in publication bias. Third, there were not sufficient data about potential interaction among TERT and MGMT, 1p/19q co-deletion, etc., If those data were adequate enough, we may achieve a better result with lower heterogeneity and clearer understanding of how pTERT mutation affects the prognosis of glioma patients.
In general, this meta-analysis demonstrated that hTERT might be the negative prognostic factors for glioma patients. The analysis of hTERT undertaken here has extended our knowledge of the impact of TERT in shortening OS were weaker on expression level than gene level. This could indicate multiple unclear mechanisms in hTERT functioning in prognosis of glioma patients. In future, more well-designed studies with different perspective are needed to confirm the conclusion. One source of weakness in this study was high heterogeneity and less effectivity of the data, which could have affected the measurements of TERT mutation and results in shorter survival time in glioma patients. Therefore, the accuracy and prospection of such predictions might be reduced. Notwithstanding these limitations, this work offers valuable insights into the diagnosis and treatment of glioma in the clinical strategy.
CONCLUSION
Although this study focuses on hTERT in glioma patients’ prognosis, one of the more significant findings to emerge from this summary is that the underling mechanism of IDH mutation reverses the poor prognosis outcome along with upregulated hTERT expression. These findings contribute in several ways to our understanding of hTERT and provide a basis for an extremely poor prognosis. Notwithstanding the relatively limited sample, this work offers valuable insights into the mechanism of hTERT expressing pathway and other involved molecules in glioma prognosis.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We are especially grateful to the National Natural Science Foundation of China Grant (No. 82174056 JD Xu; 82173795 WW) for this research. We would also like to thank Dr. Tao Xin, Department of Applied Linguistics, Capital Medical University, for polishing the language of this article.
REFERENCES
- 1.Wesseling P, Capper D. WHO 2016 Classification of gliomas. Neuropathol Appl Neurobiol. 2018;44:139–50. doi: 10.1111/nan.12432. [DOI] [PubMed] [Google Scholar]
- 2.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–50. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 3.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat Rev Genet. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 4.Powter B, Jeffreys SA, Sareen H, Cooper A, Brungs D, Po J, et al. Human TERT promoter mutations as a prognostic biomarker in glioma. J Cancer Res Clin Oncol. 2021;147:1007–17. doi: 10.1007/s00432-021-03536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo CK, Mertz D, Loeb M. Newcastle-Ottawa Scale: Comparing reviewers’ to authors’ assessments. BMC Med Res Methodol. 2014;14:45. doi: 10.1186/1471-2288-14-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao QT, Yuan Z, Zhang H, Zhang XP, Wang HE, Wang ZK, et al. Prognostic role of platelet to lymphocyte ratio in non-small cell lung cancers: A meta-analysis including 3,720 patients. Int J Cancer. 2016;139:164–70. doi: 10.1002/ijc.30060. [DOI] [PubMed] [Google Scholar]
- 7.Gramatzki D, Felsberg J, Hentschel B, Wolter M, Schackert G, Westphal M, et al. Telomerase reverse transcriptase promoter mutation- and O(6)-methylguanine DNA methyltransferase promoter methylation-mediated sensitivity to temozolomide in isocitrate dehydrogenase-wild-type glioblastoma: Is there a link? Eur J Cancer. 2021;147:84–94. doi: 10.1016/j.ejca.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Razis E, Kotoula V, Koliou GA, Papadopoulou K, Vrettou E, Giannoulatou E, et al. Is there an independent role of TERT and NF1 in high grade gliomas? Transl Oncol. 2020;13:346–54. doi: 10.1016/j.tranon.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng ZJ, Cai HQ, Zhang MJ, Zhong Y, He J, Yuan Q, et al. High S phase kinase-associated protein 2 expression is a potential prognostic biomarker for glioma. Oncol Lett. 2020;20:2788–96. doi: 10.3892/ol.2020.11818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kessler T, Berberich A, Sadik A, Sahm F, Gorlia T, Meisner C, et al. Methylome analyses of three glioblastoma cohorts reveal chemotherapy sensitivity markers within DDR genes. Cancer Med. 2020;9:8373–85. doi: 10.1002/cam4.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamrakar S, Yashiro M, Kawashima T, Uda T, Terakawa Y, Kuwae Y, et al. Clinicopathological significance of autophagy-related proteins and its association with genetic alterations in gliomas. Anticancer Res. 2019;39:1233–42. doi: 10.21873/anticanres.13233. [DOI] [PubMed] [Google Scholar]
- 12.Kuwahara K, Ohba S, Nakae S, Hattori N, Pareira ES, Yamada S, et al. Clinical, histopathological, and molecular analyses of IDH-wild-type WHO grade II-III gliomas to establish genetic predictors of poor prognosis. Brain Tumor Pathol. 2019;36:135–43. doi: 10.1007/s10014-019-00348-9. [DOI] [PubMed] [Google Scholar]
- 13.Kim HS, Kwon MJ, Song JH, Kim ES, Kim HY, Min KW. Clinical implications of TERT promoter mutation on IDH mutation and MGMT promoter methylation in diffuse gliomas. Pathol Res Pract. 2018;214:881–8. doi: 10.1016/j.prp.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen HN, Lie A, Li T, Chowdhury R, Liu F, Ozer B, et al. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 2017;19:394–404. doi: 10.1093/neuonc/now189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Wei J, Liu Y, Li P, Fan L, Wang Y, et al. Primary astrocytic tumours and paired recurrences have similar biological features in IDH1, TP53 and TERTp mutation and MGMT, ATRX loss. Sci Rep. 2017;7:13038. doi: 10.1038/s41598-017-13272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Koh J, Kim SI, Won JK, Park CK, Choi SH, et al. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol Commun. 2017;5:62. doi: 10.1186/s40478-017-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nencha U, Rahimian A, Giry M, Sechi A, Mokhtari K, Polivka M, et al. TERT promoter mutations and rs2853669 polymorphism: Prognostic impact and interactions with common alterations in glioblastomas. J Neurooncol. 2016;126:441–6. doi: 10.1007/s11060-015-1999-3. [DOI] [PubMed] [Google Scholar]
- 18.Gao K, Li G, Qu Y, Wang M, Cui B, Ji M, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7:8712–25. doi: 10.18632/oncotarget.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang ZY, Chan AK, Ding XJ, Qin ZY, Hong CS, Chen LC, et al. TERT promoter mutations contribute to IDH mutations in predicting differential responses to adjuvant therapies in WHO grade II and III diffuse gliomas. Oncotarget. 2015;6:24871–83. doi: 10.18632/oncotarget.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiegl-Kreinecker S, Lötsch D, Ghanim B, Pirker C, Mohr T, Laaber M, et al. Prognostic quality of activating TERT promoter mutations in glioblastoma: Interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro Oncol. 2015;17:1231–40. doi: 10.1093/neuonc/nov010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon M, Hosen I, Gousias K, Rachakonda S, Heidenreich B, Gessi M, et al. TERT promoter mutations: A novel independent prognostic factor in primary glioblastomas. Neuro Oncol. 2015;17:45–52. doi: 10.1093/neuonc/nou158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosrati MA, Malmström A, Lysiak M, Krysztofiak A, Hallbeck M, Milos P, et al. TERT promoter mutations and polymorphisms as prognostic factors in primary glioblastoma. Oncotarget. 2015;6:16663–73. doi: 10.18632/oncotarget.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan AK, Yao Y, Zhang Z, Chung NY, Liu JS, Li KK, et al. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod Pathol. 2015;28:177–86. doi: 10.1038/modpathol.2014.94. [DOI] [PubMed] [Google Scholar]
- 24.Labussière M, Di Stefano AL, Gleize V, Boisselier B, Giry M, Mangesius S, et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111:2024–32. doi: 10.1038/bjc.2014.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killela PJ, Pirozzi CJ, Healy P, Reitman ZJ, Lipp E, Rasheed BA, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5:1515–25. doi: 10.18632/oncotarget.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Han S, Meng L, Li Z, Zhang X, Wu A. TERT promoter mutations lead to high transcriptional activity under hypoxia and temozolomide treatment and predict poor prognosis in gliomas. PLoS One. 2014;9:e100297. doi: 10.1371/journal.pone.0100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126:931–7. doi: 10.1007/s00401-013-1163-0. [DOI] [PubMed] [Google Scholar]
- 28.Gandhi P, Shrivastava R, Garg N, Sorte SK. Novel molecular panel for evaluating systemic inflammation and survival in therapy naïve glioma patients. World J Clin Oncol. 2021;12:947–59. doi: 10.5306/wjco.v12.i10.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elsers D, Temerik DF, Attia AM, Hadia A, Hussien MT. Prognostic role of ALK-1 and h-TERT expression in glioblastoma multiforme: Correlation with ALK gene alterations. J Pathol Transl Med. 2021;55:212–24. doi: 10.4132/jptm.2021.03.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potharaju M, Mathavan A, Mangaleswaran B, Patil S, John R, Ghosh S, et al. Clinicopathological analysis of HIF-1alpha and TERT on survival outcome in glioblastoma patients: A prospective, single institution study. J Cancer. 2019;10:2397–406. doi: 10.7150/jca.32909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rute JM, Martínez M, Gibert J, Navarro P, Bellosillo B, Alameda F. Prognostic value of tumoral stem cells in glioblastoma: Immunohistochemical study of CD133 and HTERT expression. Anal Quant Cytopathol Histopathol. 2017;39:259–64. [Google Scholar]
- 32.Gandhi P, Khare R, Garg N. Evaluating the potential of circulating hTERT levels in glioma: Can plasma levels serve as an independent prognostic marker? J Neurooncol. 2017;135:255–61. doi: 10.1007/s11060-017-2578-6. [DOI] [PubMed] [Google Scholar]
- 33.Dorris K, Sobo M, Onar-Thomas A, Panditharatna E, Stevenson CB, Gardner SL, et al. Prognostic significance of telomere maintenance mechanisms in pediatric high-grade gliomas. J Neurooncol. 2014;117:67–76. doi: 10.1007/s11060-014-1374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lötsch D, Ghanim B, Laaber M, Wurm G, Weis S, Lenz S, et al. Prognostic significance of telomerase-associated parameters in glioblastoma: Effect of patient age. Neuro Oncol. 2013;15:423–32. doi: 10.1093/neuonc/nos329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahrami A, Lee S, Schaefer IM, Boland JM, Patton KT, Pounds S, et al. TERT promoter mutations and prognosis in solitary fibrous tumor. Mod Pathol. 2016;29:1511–22. doi: 10.1038/modpathol.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 37.Vogazianou AP, Chan R, Bäcklund LM, Pearson DM, Liu L, Langford CF, et al. Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses. Neuro Oncol. 2010;12:664–78. doi: 10.1093/neuonc/nop075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–97. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]