Figure 3: RNA binding proteins and regulation by titrating sRNAs.
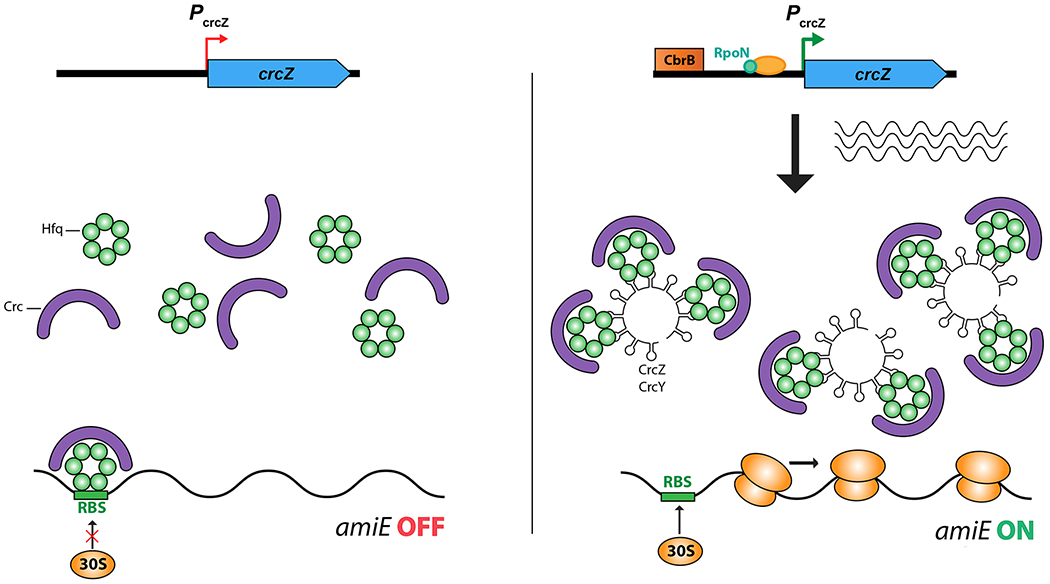
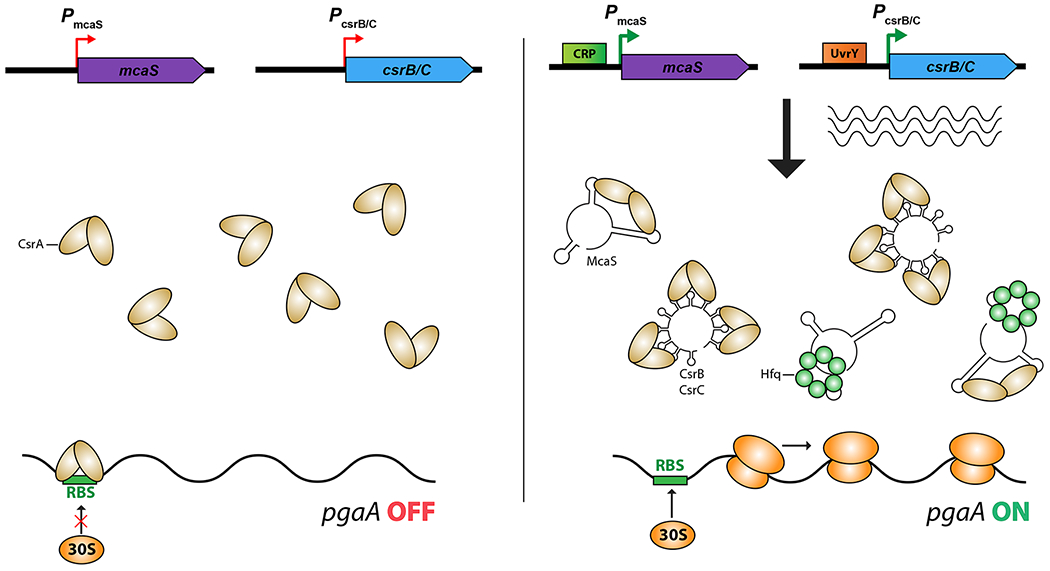
RNA binding proteins can be removed from their regulatory sites on target mRNAs by competing RNAs that also carry the regulatory sites. These titrating RNAs may be the major regulatory input affecting translational regulatory proteins.
A. Titration of CsrA: CsrA is a major regulatory protein that acts by binding to targets, frequently blocking ribosome access (reviewed in [32]). All bacteria that contain CsrA or its homologs also encode titrating sRNAs such as CsrB and CsrC, which each contain multiple CsrA binding sites. Recent work demonstrates that some Hfq-binding sRNAs that act by pairing, such as McaS, also contain CsrA binding sites and thus can titrate CsrA as well [30,33]. Different regulatory signals affect synthesis of different titrating sRNAs.
B. In Pseudomonads, Hfq acts in concert with another RNA binding protein, Crc, to carry out sRNA-independent catabolite repression at multiple sites. The CrcZ regulatory RNA, when synthesized, acts to remove Hfq and Crc from targets, allowing expression of genes such as amiE (shown here) [52]. The expression of CrcZ is dependent upon a response regulator, CbrB, which responds to the status of carbon sources to set the hierarchy of carbon utilization [51].
