Fig. 3 |. Structural basis of high-mannose N-glycan recognition by Bl_Man38A–Bl_Man38C.
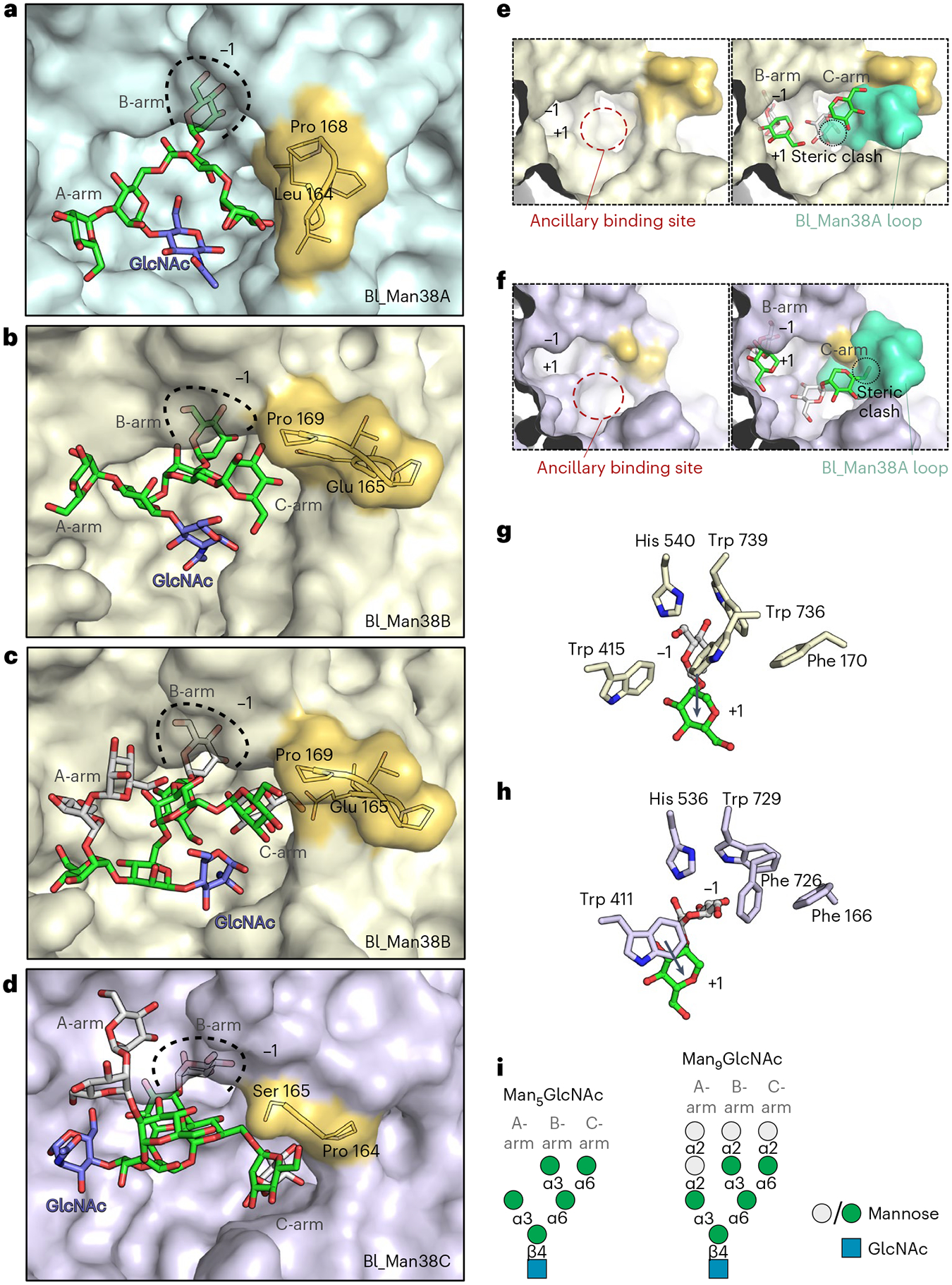
a–d, Representative structures of Bl_Man38A (a), Bl_Man38B (b and c) and Bl_Man38C (d) after docking and energy minimization with Man5GlcNAc (a and b) and Man9GlcNAc (c and d). e,f, Superposition of the hypervariable loop of Bl_Man38A (green) to the active sites of Bl_Man38B (e) and Bl_Man38C (f), showing only the B- and C-arms of Man9GlcNAc (docked and energy-minimized complexes). g,h, Comparison of the putative interactions formed by Trp 736 in Bl_Man38B (g) and its substitution for a phenylalanine residue in Bl_Man38C (Phe 726) (h), showing the shift of orientation of the α-1,2-mannoside in the active site of the enzymes. Only the α-1,2-mannoside of Man9GlcNAc is shown for better visualization. i, Schematic representation of Man5GlcNAc and Man9GlcNAc docked into the structures.
