Extended Data Fig. 1 |. Genomic organization of the predicted N-glycan utilization loci (N-GUL) from B. longum NCC2705.
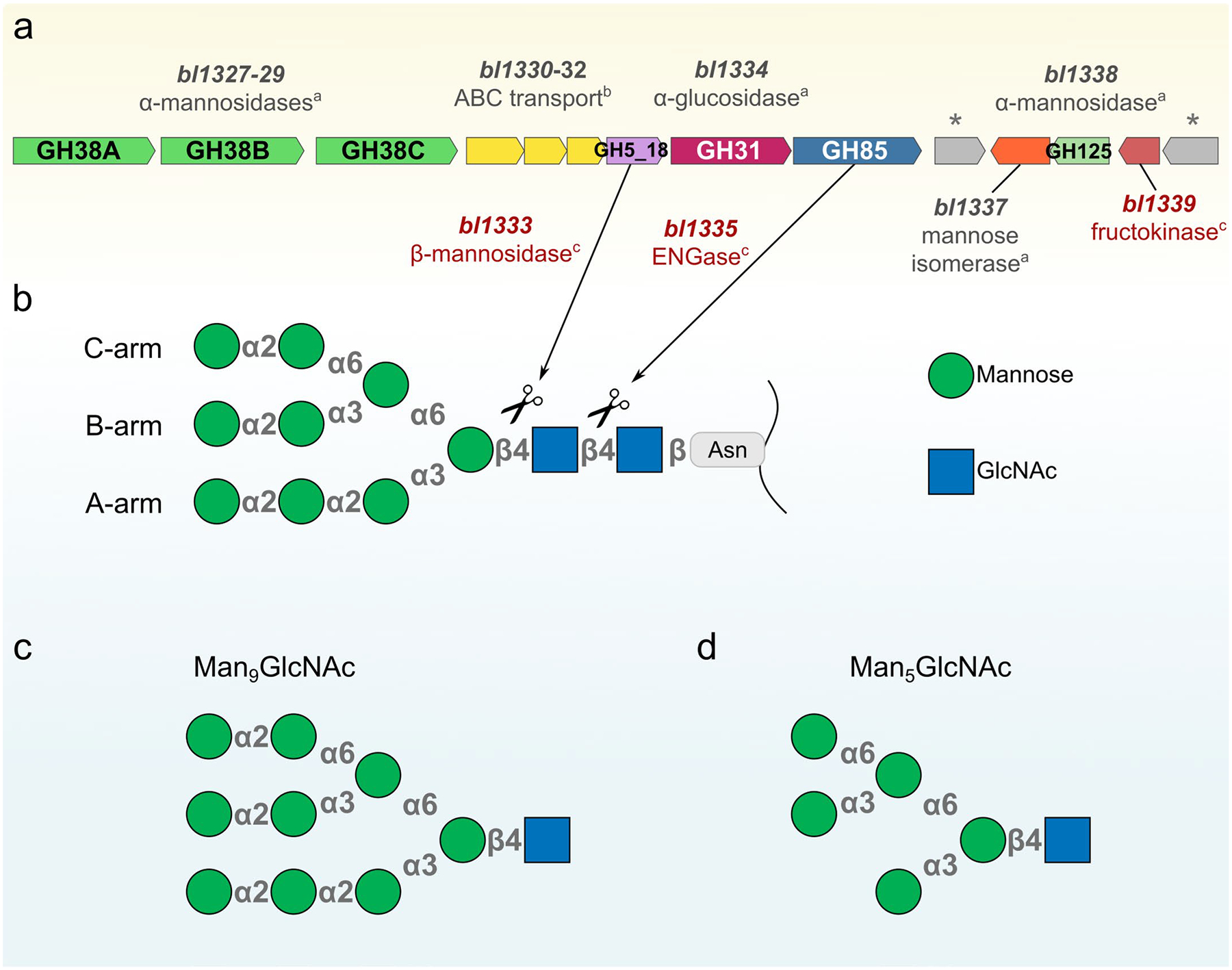
a, Genes that compose N-GUL represented by arrows and their respective gene products. a = biochemically characterized in this work, b = predicted, c = characterized in previous works23,24,28. The symbol * represents transcriptional regulators (bl1336 – encoding a LacI transcriptional regulator and bl1340 – encoding a ROK transcriptional regulator). b, A schematic representation of a high-mannose N-glycan indicating its subunits (geometric forms), glycosidic linkages (gray symbols) and the cleavage sites (scissors) recognized by the GH5 β-mannosidase23 and GH85 ENGase24 enzymes previously characterized. Predictions of uncharacterized enzymes were performed by sequence and hidden-Markov similarities analyses (Supplementary Tables 1, 2). c, Representation of Man9GlcNAc. d, Representation of Man5GlcNAc.
