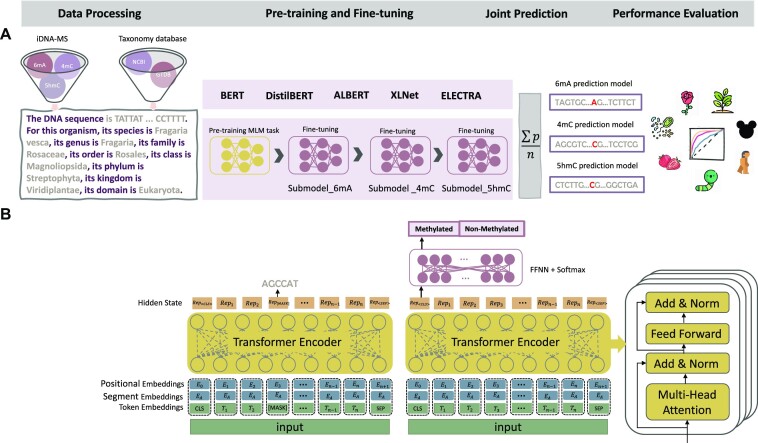
Figure 1:
The MuLan-Methyl workflow. (A) The framework employs 5 fine-tuned language models for joint identification of DNA methylation sites. Methylation datasets (obtained from iDNA-MS) were processed as sentences that describe the DNA sequence as well as the taxonomy lineage, giving rise to the processed training dataset and the processed independent set. For each transformer-based language model, a custom tokenizer was trained based on a corpus of the processed training dataset and taxonomy lineage data. Pretraining and fine-tuning were both conducted on each methylation site-specific training subset separately. During model testing, the prediction of a sample in the processed independent test set is defined as the average prediction probability of the 5 fine-tuned models. We thus obtained 3 methylation type-wise prediction models. We evaluated the model performance on the genome type that contained the corresponding methylation type-wise dataset, respectively. In total, we evaluated 17 combinations of methylation types and taxonomic lineages. (B) The general transformer-based language model architecture for pretraining and fine-tuning. The model was pretrained using the masked language modeling (MLM) task and then fine-tuned on the methylation type-wise processed training dataset.